Brief Overview
Overview of Genomics
Genomics is the study of all of a person’s genes (the genome), including interactions of those genes with each other and with the person’s environment. Genome is an organisms complete set of DNA, which itself is composed of 4 letters (base bairs) A, G, T, C. The word genome was first coined by Tom Roderick, a geneticist at Jackson laboratory in 1986. While ‘Genetics’ is the study of genes and its inheritance, Genomics is the study of the entire genome including structure, function and interactions. 1 Each human genome is around 3.2 billions letters (A,G,C,T) of your DNA code and three letters combinations forms the codon and each codon specifies a particular amino acid. The amino acids provide instructions that can be read during the process of translation to form a protein which is a chain of amino acid sequences. Genes are the instructions for making the proteins and it forms around 1.5% of your genome. The rest of the region between genes is called ‘junk DNA’ which was discovered to regulate genes and the genome. Genomics studies this entire genome including genes and ‘junk DNA’
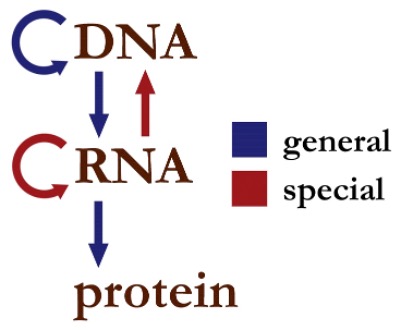 image credit: Wikipedia
image credit: Wikipedia
Central Dogma of Molecular Biology
The Central Dogma of Molecular biology is the process of flow of genetic information from DNA to functional products first defined by Francis Crick (discovered structure of DNA) in 1958. It states that genetic information flows from DNA in your cells which is converted into RNA through a process called transcription. During the second phase of translation, these RNA travel from the cells to the ribosomes and are then translated into proteins (the functional product). The pattern of information that occurs in the cells are: 1. DNA to DNA (Replication), DNA to RNA (transcription) and RNA to Protein (Translation)
Databases
The number of database resources that manage and store sequence data are also growing. The National Center for Biotechnology Information, European Molecular Biology Laboratory (EMBL) and the DNA Databank of Japan(DDBJ) together have created the International Nucleotide Sequence Database collaboration to store, organize and distribute nucleotide and amino acid sequences that become available. The challenge with the use of these databases is that these repositories are not annotated and organized in a way that can be used by researchers. To answer these challenges a number of general purpose genome browsers like the University of California, Santacruz (UCSC) Genome Browser, EBI’s Ensemble and NCBI’s MapViewer that provide a more genomic context for genome level features like genes and disease loci.
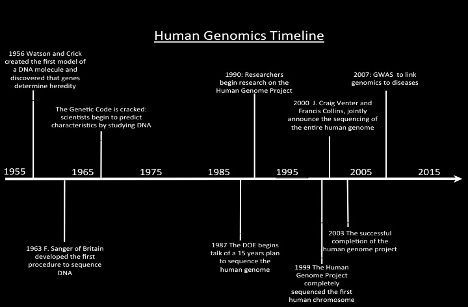 image credit: O’Reilly
image credit: O’Reilly
The Human Genome Project
Began formally in 1990, The Human Genome project was an ambitious 13 year effort between National Institutes of Health and Department of Energy to determine the sequence of the entire human genome. The HGP had revealed that there are probably about 20,500 human genes. This ultimate product of the HGP has given the world a resource of detailed information about the structure, organization and function of the complete set of human genes. This information can be thought of as the basic set of inheritable “instructions” for the development and function of a human being2. This project mapped about 92% of the human genome which was immediately made public and served as a great tool for researchers to study the whole genome.
The last 8% of the genome mapping was recently completed and published by the Telomere to Telomere (T2T) consortium. 3 The researchers generated this using DNA from Human cell line which were mostly DNA sequences near the repetitive telomere (the ends portion of each chromosome) and centromeres (the middle portion of each chromosome).
Subfields within Genomics are:
-
Comparative Genomics: This is the study of the comparision of genomes of different organanism or the genomes of strains within a species. It is a great tool to study evolution.
-
Structural Genomics: Structural genomics is branch of structural biology that refers to the exploration and determination of three dimensional protein structures on a genome scale.
-
Functional Genomics: Functional Genomics is the field that studies the functions of genes and its interactions to determine the relationship between the organism’s genome
and its phenotype. DNA: Genomics and epigenomics RNA: transcriptomics Protein: Proteomics Metabolite: Metabolomics -
Epigenomics: The study involving the investigation of interactions between genes and between genes and the environment.
Challenges:
The genomics data has gone through an exponential growth with the decreasing cost of sequencing data since the completion of the Human Genome Project in 2000. With the growing interest in Genomics research, the field has faced several technical and data privacy and security challenges.
Infrastructure
As pieces dropped for sequencing over the years, even small labs have been generating big data. This resulted in labs including small research groups generating large datasets on patient samples and model organisms. This puts the burden of storing, managing, analyzing and interpreting the data on the scientists who might not be very computer savvy. Often these projects also are a part of collaboartions between groups making data sharing and transfer a challenge. Comparing human genomes takes more than a personal computer and online file-sharing applications. This requires researchers to have hardware’s that often require maintenance and comes with an upfront cost. Many of the large-scale human genomics datasets have been made public and are available to download on demand. Downloading such large datasets on local or personal computers is not an easy task. An alternative to this is using cloud for both computing and storage where researchers can bring their tools to the data. This eliminates the need to move data around and the analyses can be performed right in the cloud with approprite resources for the task. The pay-as-you-go model of cloud computing doesnot put any upfront cost for the researchers and only charges for the resources being used. [4,5]
The benefit of cloud comes from its elasticity and scalability on demand. There is another benefit of cloud with its global accessibility where a researcher can access the resources from anywhere in the world.
The benefit of cloud comes from its elasticity and scalability on demand. There is another benefit of cloud with its global accessibility where a researcher can access the resources from anywhere in the world.
Reproducibility:
FAIR data
Data sharing and re-use of data are the prerequisites to achieving the benefits of the data driven approach in genomics research. FAIR principles have garnered a lot of attention since its inception in 2014 as a set of minimal guiding principles for data stewardship in life sciences. Various initiatives and organization are working together towards setting guidelines in making data more Findable, Accessible, Interoperable and Re-usable. The use of unique global identifier, accompanying rich metadata and indexed by a system for easy retrieval makes data findable. Accessibility requires use of standardized, open communications protocol for retrieval of data by the unique identifier. Interoperability is supported using standardized and machine actionable metadata and data. In order to make data re-usable, the accompanying metadata is well described with standard vocabulary, contains information on provenance and has a clear data usage license with it. 6
Table 7
| Principle | Explanation | Example in human genetics and genomics |
|---|---|---|
| Findability | Datasets should be described, identified and registered or indexed in a clear and unequivocal manner | BBMRI-ERIC Directory |
| Accessibility | Datasets should be accessible through a clearly defined access procedure, ideally using automated means. Metadata should always remain accessible | European genome–phenome archive |
| Interoperability | Data and metadata are conceptualised, expressed and structured using common, published standards | GA4GH Genomic Data Toolkit |
| Reusability | Characteristics of data and their provenance are described in detail according to domain- relevant community standards, with clear and accessible conditions for use | BRCA exchange |
Genomics in Clinic
The technological advance and lowering cost of re-sequencing of patient samples have led to an expansion of clinical testing for Mendelian diseases and complex diseases. Large scale sequencing programs like The Cancer Genome Atlas, TCGA, gnomAD and1UK10K havehelped in the interpretation of variants. However there are still certain challenges facing adoption of next generation sequencing in clinical practice including standards for reporting data, incidental findings, data storage, patient privacy, CLIA/ISO certification. Moreover, the technologies and computational methods used need to be validated against required metrics using benchmarking datasets like the Genome In A Bottle (GIAB) and the Platinum Genome
References and further reading: