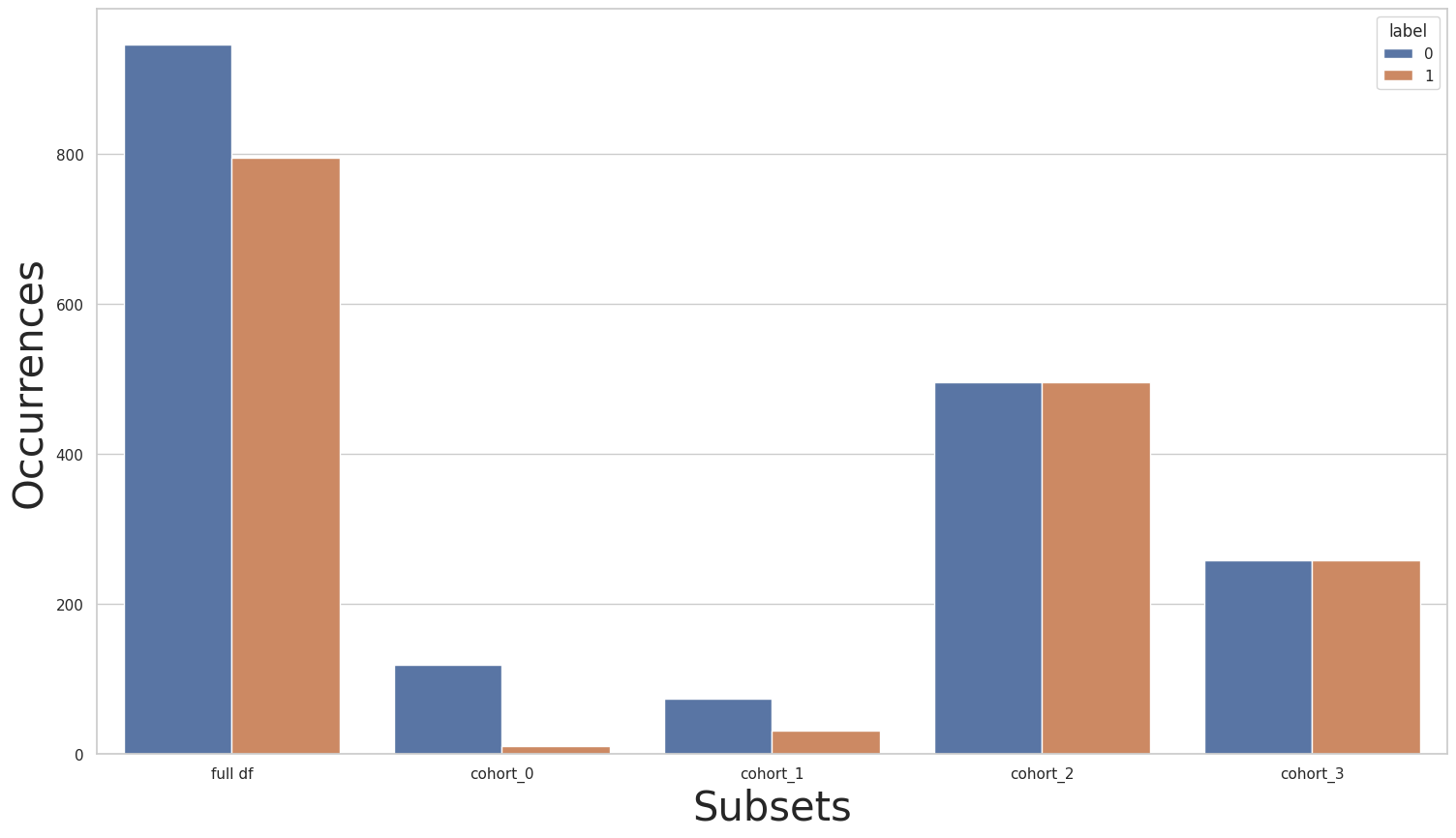
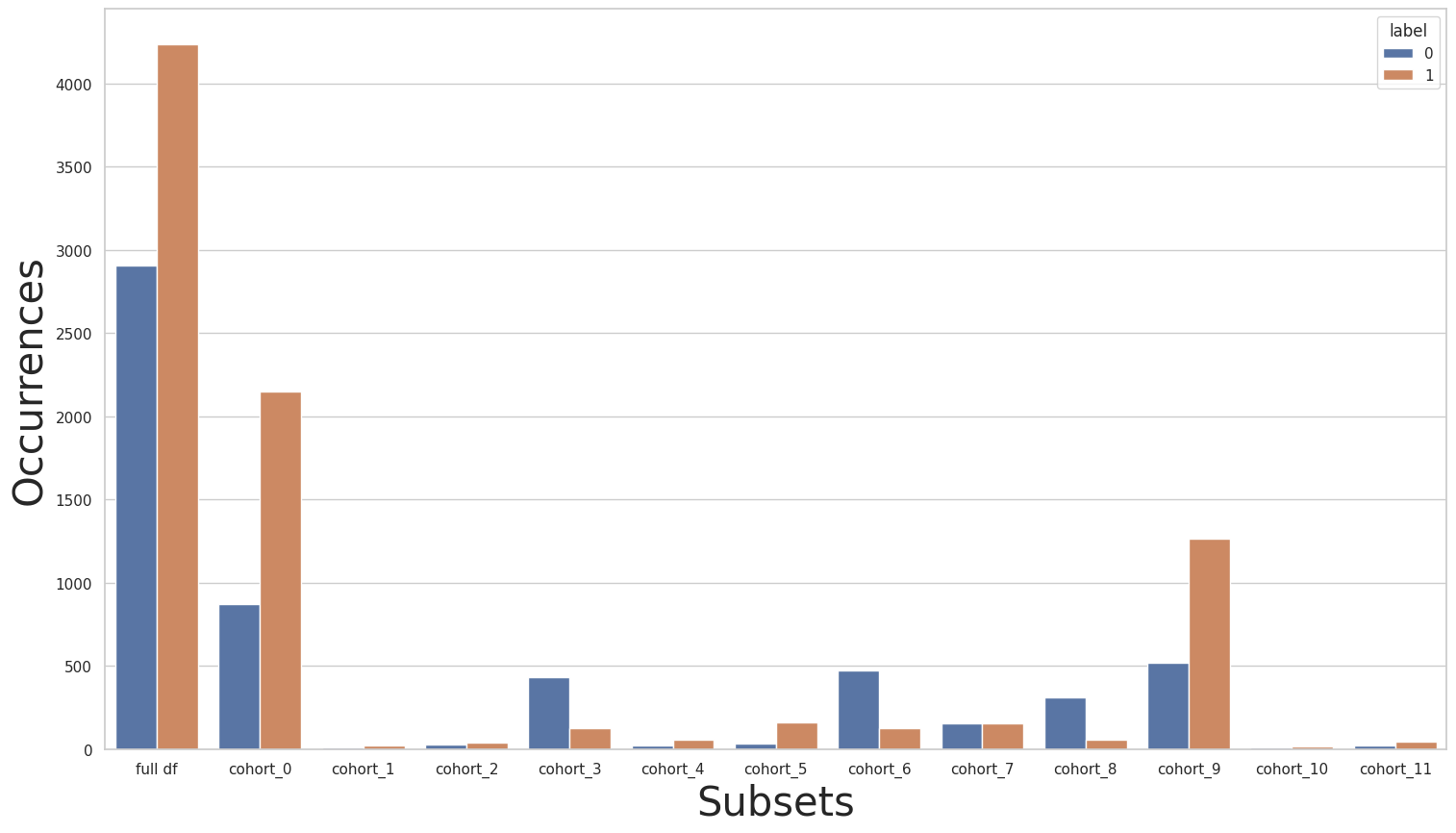
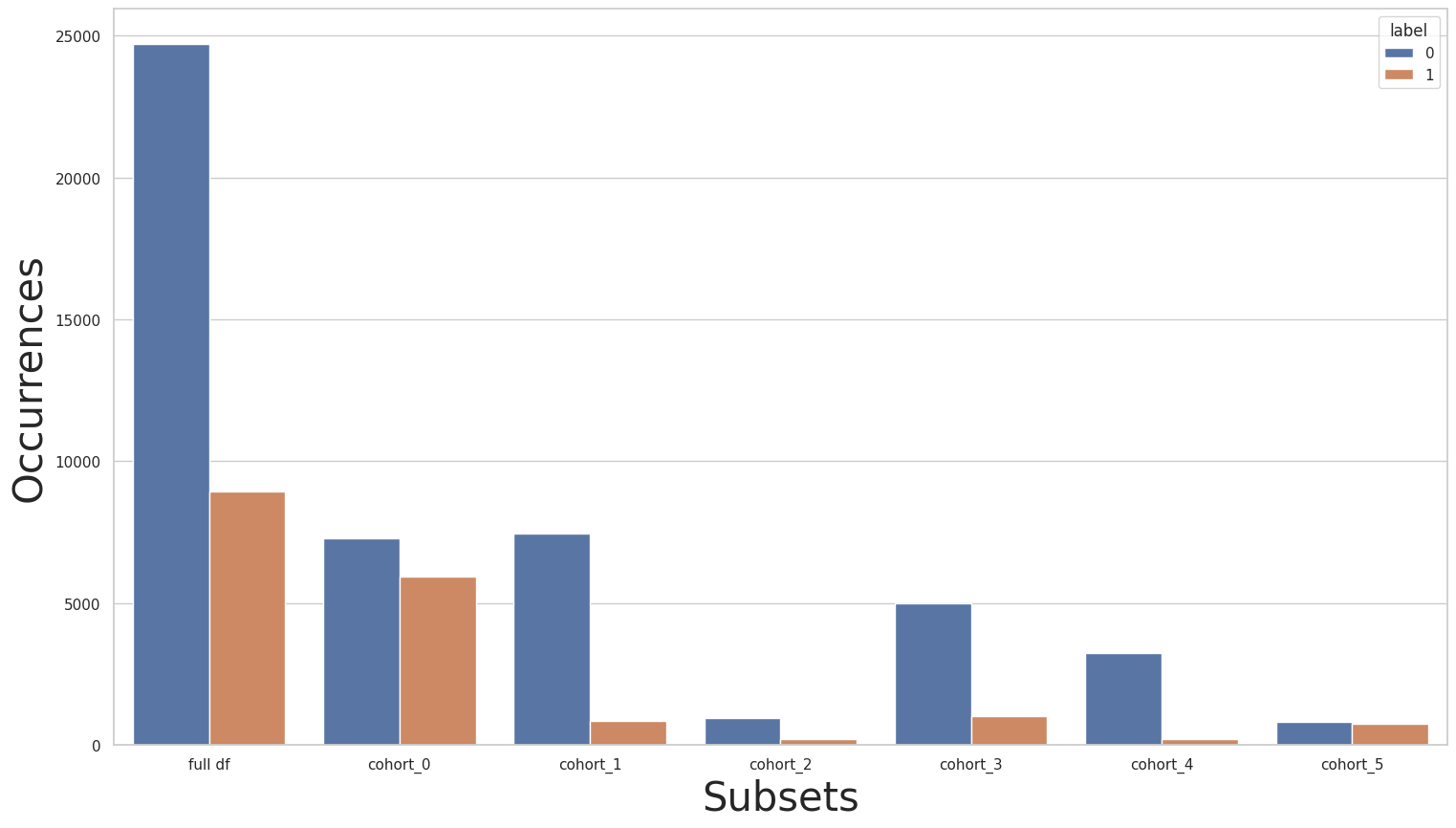
CohortManager
The CohortManager allows the application of different data processing pipelines over each cohort. Also allows the creation and
filtering of multiple cohorts using a simple interface. Finally, allows the creation of different estimators for each cohort using
the .predict() and predict_proba() interfaces. This class uses the cohort.CohortDefinition
internally in order to create, filter, and manipulate multiple cohorts. There are multiple ways of using the
cohort.CohortManager class when building a pipeline, and these different scenarios are summarized in following
figure.
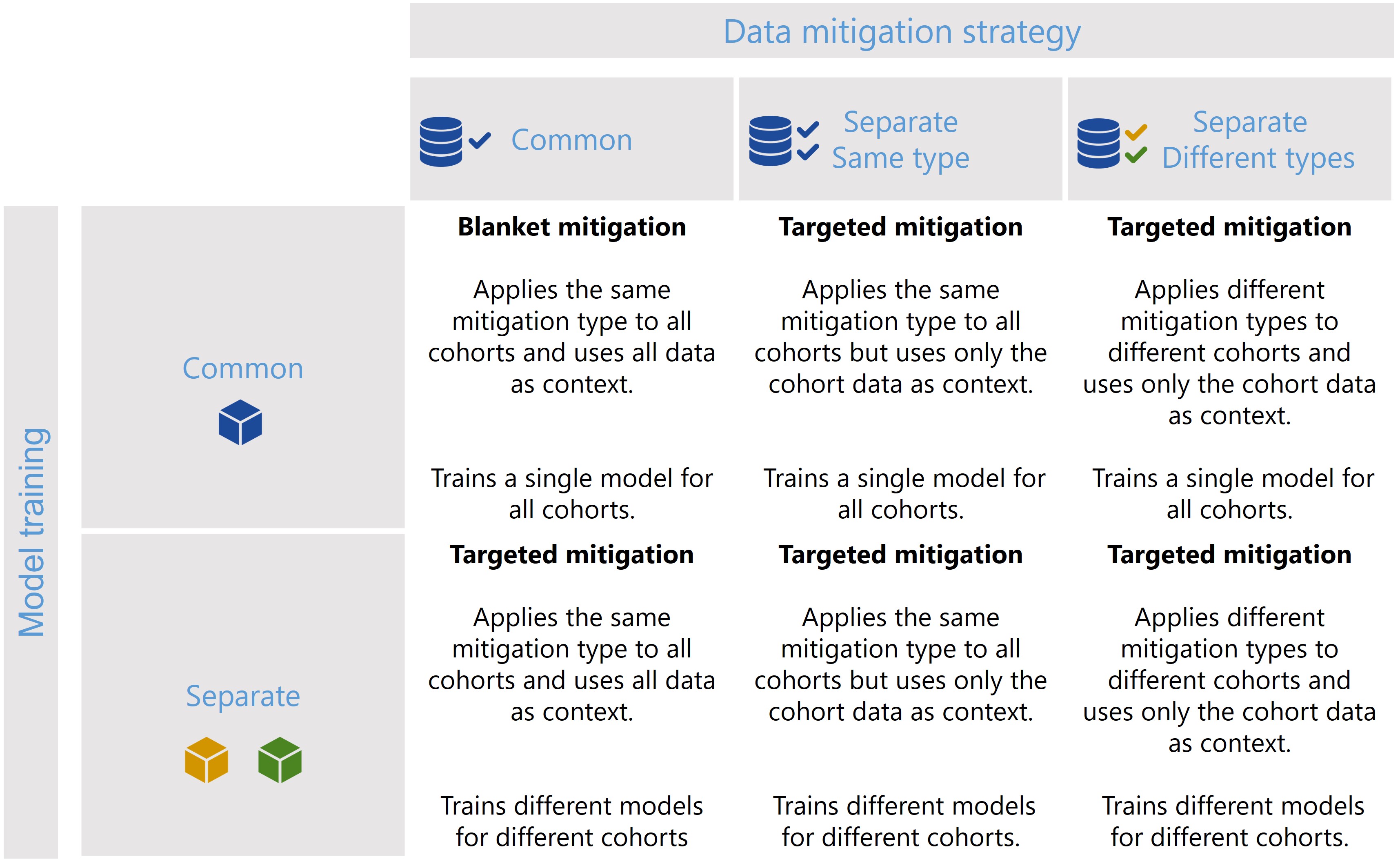
Figure 1 - The CohortManager class can be used in different ways to target mitigations to different cohorts. The main differences between these scenarios consist on whether the same or different type of data mitigation is applied to the cohort data, and whether a single or separate models will be trained for different cohorts. Depending on these choices, CohortManager will take care of slicing the data accordingly, applying the specified data mitigation strategy, merging the data back, and retraining the model(s).
The Cohort Manager - Scenarios and Examples notebook, located in notebooks/cohort/cohort_manager_scenarios.ipynb and listed in
the Examples section below, shows how each of these scenarios can be implemented through simple code snippets.
- class raimitigations.cohort.CohortManager(transform_pipe: Optional[list] = None, cohort_def: Optional[Union[dict, list, str]] = None, cohort_col: Optional[list] = None, cohort_json_files: Optional[list] = None, df: Optional[DataFrame] = None, label_col: Optional[str] = None, X: Optional[DataFrame] = None, y: Optional[DataFrame] = None, verbose: bool = True)
Concrete class that manages multiple cohort pipelines that are applied using the
fit(),transform(),fit_resample(),predict(), andpredict_proba()interfaces. TheCohortManageruses multipleCohortDefinitionobjects to control the filters of each cohort, while using transformation pipelines to control which transformations should be applied to each cohort.- Parameters
transform_pipe –
the transformation pipeline to be used for each cohort. There are different ways to present this parameter:
An empty list or None: in this case, the
CohortManagerwon’t apply any transformations over the dataset. Thetransform()method will simply return the dataset provided;A single transformer: in this case, this single transformer is placed in a list (a list with a single transformer), which is then replicated such that each cohort has its own list of transformations (pipeline);
A list of transformers: in this case, this pipeline is replicated for each cohort;
A list of pipelines: a list of pipelines is basically a list of lists of transformations. In this case, the list of pipelines should have one pipeline for each cohort created, that is, the length of the
transform_pipeparameter should be the same as the number of cohorts created. The pipelines will be assigned to each cohort following the same order as thecohort_defparameter (depicted in the following example);
cohort_def – a list of cohort definitions or a dictionary of cohort definitions. A cohort condition is the same variable received by the
cohort_definitionparameter of theCohortDefinitionclass. When using a list of cohort definitions, the cohorts will be named automatically. For the dictionary of cohort definitions, the key used represents the cohort’s name, and the value assigned to each key is given by that cohort’s conditions. This parameter can’t be used together with thecohort_colparameter. Only one these two parameters must be used at a time. This parameter is ignored ifcohort_json_filesis provided;cohort_col – a list of column names or indices, from which one cohort is created for each unique combination of values for these columns. This parameter can’t be used together with the
cohort_defparameter. Only one these two parameters must be used at a time. This parameter is ignored ifcohort_json_filesis provided;cohort_json_files – a list with the name of the JSON files that contains the definition of each cohort. Each cohort is saved in a single JSON file, so the length of the
cohort_json_filesshould be equal to the number of cohorts to be used.df – the data frame to be used during the fit method. This data frame must contain all the features, including the label column (specified in the
label_colparameter). This parameter is mandatory iflabel_colis also provided. The user can also provide this dataset (along with thelabel_col) when calling thefit()method. If df is provided during the class instantiation, it is not necessary to provide it again when callingfit(). It is also possible to use theXandyinstead ofdfandlabel_col, although it is mandatory to pass the pair of parameters (X,y) or (df, label_col) either during the class instantiation or during thefit()method;label_col – the name or index of the label column. This parameter is mandatory if
dfis provided;X – contains only the features of the original dataset, that is, does not contain the label column. This is useful if the user has already separated the features from the label column prior to calling this class. This parameter is mandatory if
yis provided;y – contains only the label column of the original dataset. This parameter is mandatory if
Xis provided;verbose – indicates whether internal messages should be printed or not.
- fit(X: Optional[Union[DataFrame, ndarray]] = None, y: Optional[Union[Series, ndarray]] = None, df: Optional[DataFrame] = None, label_col: Optional[str] = None)
Calls the
fit()method of all transformers in all pipelines. Each cohort has its own pipeline. This way, the following steps are executed: (i) iterate over each cohort, (ii) filter the dataset (Xordf) using each cohort’s filter, (iii) cycle through each of the transformers in the cohort’s pipeline and call the transformer’sfit()method, (iv) after fitting the transformer, call itstransform()method to get the updated subset, which is then used in thefit()call of the following transformer. Finally, check if all instances belong to only a single cohort.- Parameters
X – contains only the features of the original dataset, that is, does not contain the label column;
y – contains only the label column of the original dataset;
df – the full dataset;
label_col – the name or index of the label column;
Check the documentation of the _set_df_mult method (DataProcessing class) for more information on how these parameters work.
- fit_resample(X: Optional[Union[DataFrame, ndarray]] = None, y: Optional[Union[DataFrame, ndarray]] = None, df: Optional[Union[DataFrame, ndarray]] = None, rebalance_col: Optional[str] = None)
Calls the
fit_resample()method of all transformers in all pipelines. Each cohort has its own pipeline. This way, the following steps are executed: (i) iterate over each cohort, (ii) filter the dataset (Xordf) using each cohort’s filter, (iii) cycle through each of the transformers in the cohort’s pipeline and call the transformer’sfit_resample()method, (iv) after resampling using the current transformer, save the new subset and use it when calling thefit_resample()of the following transformer. Finally, check if all instances belong to only a single cohort.- Parameters
X – contains only the features of the original dataset, that is, does not contain the column used for rebalancing. This is useful if the user has already separated the features from the label column prior to calling this class. This parameter is mandatory if
yis provided;y – contains only the rebalance column of the original dataset. The rebalance operation is executed based on the data distribution of this column. This parameter is mandatory if
Xis provided;df – the dataset to be rebalanced, which is used during the
fit()method. This data frame must contain all the features, including the rebalance column (specified in therebalance_colparameter). This parameter is mandatory ifrebalance_colis also provided. The user can also provide this dataset (along with therebalance_col) when calling thefit()method. Ifdfis provided during the class instantiation, it is not necessary to provide it again when callingfit(). It is also possible to use theXandyinstead ofdfandrebalance_col, although it is mandatory to pass the pair of parameters (X,y) or (df, rebalance_col) either during the class instantiation or during thefit()method;rebalance_col – the name or index of the column used to do the rebalance operation. This parameter is mandatory if
dfis provided.
- Returns
the resampled dataset.
- Return type
pd.DataFrame
- predict(X: Union[DataFrame, ndarray], split_pred: bool = False)
Calls the
transform()method of all transformers in all pipelines, followed by thepredict()method for the estimator (which is always the last object in the pipeline).- Parameters
X – contains only the features of the dataset to be transformed;
split_pred – if True, return a dictionary with the predictions for each cohort. If False, return a single predictions array;
- Returns
an array with the predictions of all instances of the dataset, built from the predictions of each cohort, or a dictionary with the predictions for each cohort;
- Return type
np.ndarray or dict
- predict_proba(X: Union[DataFrame, ndarray], split_pred: bool = False)
Calls the
transform()method of all transformers in all pipelines, followed by thepredict_proba()method for the estimator (which is always the last object in the pipeline).- Parameters
X – contains only the features of the dataset to be transformed;
split_pred – if True, return a dictionary with the predictions for each cohort. If False, return a single predictions array;
- Returns
an array with the predictions of all instances of the dataset, built from the predictions of each cohort, or a dictionary with the predictions for each cohort;
- Return type
np.ndarray or dict
- transform(X: Union[DataFrame, ndarray])
Calls the
transform()method of all transformers in all pipelines. Each cohort has its own pipeline. This way, the following steps are executed: (i) iterate over each cohort, (ii) filter the datasetXusing each cohort’s filter, (iii) cycle through each of the transformers in the cohort’s pipeline and call the transformer’stransform()method, which returns a new transformed subset, that is then used in thetransform()call of the following transformer. Finally, check if all instances belong to only a single cohort, and merge all cohort subsets into a single dataset.- Parameters
X – contains only the features of the dataset to be transformed;
- Returns
a dataset containing the transformed instances of all cohorts.
- Return type
pd.DataFrame
Class Diagram