[13]:
import pandas as pd
import numpy as np
import uci_dataset as database
import torch
from raimitigations.utils import train_model_plot_results, split_data
import raimitigations.dataprocessing as dp
Case Study 2
Fixing a seed
To avoid randomness in the following experiments, we’ll fix the seeds to guarantee that the results obtained are the same each time we run this notebook. Feel free to comment the next cell or test different seeds to see how this affects the results.
[14]:
import random
SEED = 42
np.random.seed(SEED)
random.seed(SEED)
torch.manual_seed(SEED)
[14]:
<torch._C.Generator at 0x7ffb56dee9f0>
1 - Understanding the Data
In this example, we will build a classifier to detect diseased trees using the dataprocessing library, with data from the uci_dataset. The data itself comes from spectral imagery, and the data columns relate to features in the imagery.
The data columns have the following meaning:
class: ‘w’ (diseased trees), ‘n’ (all other land cover)
GLCM_Pan: GLCM mean texture (Pan band)
Mean_G: Mean green value
Mean_R: Mean red value
Mean_NIR: Mean NIR value
SD_Pan: Standard deviation (Pan band)
[15]:
df = database.load_wilt()
label_col = "class"
df
[15]:
| class | GLCM_pan | Mean_Green | Mean_Red | Mean_NIR | SD_pan | |
|---|---|---|---|---|---|---|
| 0 | w | 120.362774 | 205.500000 | 119.395349 | 416.581395 | 20.676318 |
| 1 | w | 124.739583 | 202.800000 | 115.333333 | 354.333333 | 16.707151 |
| 2 | w | 134.691964 | 199.285714 | 116.857143 | 477.857143 | 22.496712 |
| 3 | w | 127.946309 | 178.368421 | 92.368421 | 278.473684 | 14.977453 |
| 4 | w | 135.431548 | 197.000000 | 112.690476 | 532.952381 | 17.604193 |
| ... | ... | ... | ... | ... | ... | ... |
| 495 | n | 123.554348 | 202.826087 | 106.391304 | 364.565217 | 17.314068 |
| 496 | n | 121.549028 | 276.220000 | 175.593333 | 402.620000 | 13.394574 |
| 497 | n | 119.076687 | 247.951220 | 113.365854 | 808.024390 | 24.830059 |
| 498 | n | 107.944444 | 197.000000 | 90.000000 | 451.000000 | 8.214887 |
| 499 | n | 119.731928 | 182.238095 | 74.285714 | 301.690476 | 22.944278 |
4839 rows × 6 columns
[16]:
df[label_col] = df[label_col].replace({"w": 1, "n": 0})
df.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 4839 entries, 0 to 499
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 class 4839 non-null int64
1 GLCM_pan 4839 non-null float64
2 Mean_Green 4839 non-null float64
3 Mean_Red 4839 non-null float64
4 Mean_NIR 4839 non-null float64
5 SD_pan 4839 non-null float64
dtypes: float64(5), int64(1)
memory usage: 264.6 KB
[17]:
df.isna().any()
[17]:
class False
GLCM_pan False
Mean_Green False
Mean_Red False
Mean_NIR False
SD_pan False
dtype: bool
First, we will try to determine if any of the features are correlated, and remove the correlated features.
[18]:
cor_feat = dp.CorrelatedFeatures(
method_num_num=["spearman", "pearson", "kendall"], # Used for Numerical x Numerical correlations
num_corr_th=0.9, # Used for Numerical x Numerical correlations
num_pvalue_th=0.05, # Used for Numerical x Numerical correlations
method_num_cat="model", # Used for Numerical x Categorical correlations
model_metrics=["f1", "auc"], # Used for Numerical x Categorical correlations
metric_th=0.9, # Used for Numerical x Categorical correlations
cat_corr_th=0.9, # Used for Categorical x Categorical correlations
cat_pvalue_th=0.01, # Used for Categorical x Categorical correlations
json_summary="./corr_json/c2_summary.json",
json_corr="./corr_json/c2_corr.json",
json_uncorr="./corr_json/c2_uncorr.json"
)
cor_feat.fit(df=df, label_col=label_col)
No correlations detected. Nothing to be done here.
[18]:
<raimitigations.dataprocessing.feat_selection.correlated_features.CorrelatedFeatures at 0x7ffaf7250a60>
Remember to look through the JSON files generated in the previous cell. But, according to the thresholds we have set, no features are correlated.
2 - Baseline Models
[19]:
df[label_col].value_counts()
[19]:
0 4578
1 261
Name: class, dtype: int64
In this example, we have an imbalanced dataset (most trees are not diseased). While we will take a look at a number of different metrics, we will be focused on improved the F1 score for this dataset.
After splitting the data into train and test sets, we will build two baseline models, one with XGBoost, and the other with KNN.
[20]:
train_x, test_x, train_y, test_y = split_data(df, label_col, test_size=0.25)
[21]:
model = train_model_plot_results(train_x, train_y, test_x, test_y, model="xgb", train_result=False, plot_pr=False)
TEST SET:
[[1115 30]
[ 6 59]]
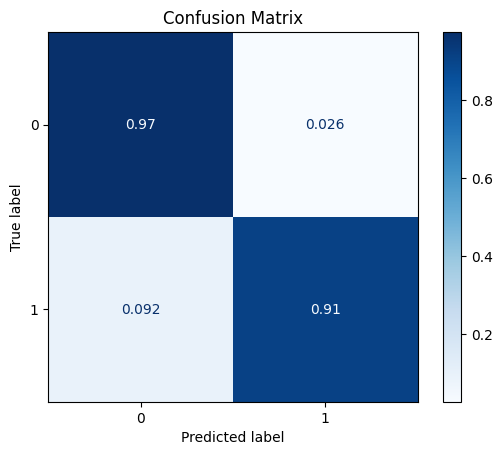
ROC AUC: 0.9681558616056434
Precision: 0.8287844921769287
Recall: 0.9407457171649312
F1: 0.8751733703190014
Accuracy: 0.9702479338842975
Optimal Threshold (ROC curve): 0.15434882044792175
Optimal Threshold (Precision x Recall curve): 0.1898476928472519
Threshold used: 0.15434882044792175
[22]:
model = train_model_plot_results(train_x, train_y, test_x, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1120 25]
[ 8 57]]
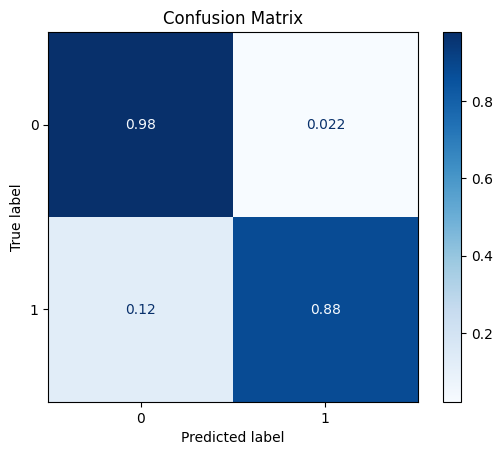
ROC AUC: 0.9348202888814242
Precision: 0.8440148763189759
Recall: 0.9275445078938529
F1: 0.8804959731362849
Accuracy: 0.9727272727272728
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
For a baseline, XGBoost and KNN have F1 scores around 0.86 (this will depend on your train/test split if you rerun this notebook). We will now proceed to try and improve on these results.
3 - Data Transformation
DataMinMaxScaler
The first data transformation we will perform is a MinMaxScaler, which will scale each feature to have a range between zero and one. This does not improve the results.
[23]:
scaler = dp.DataMinMaxScaler()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1018 127]
[ 23 42]]
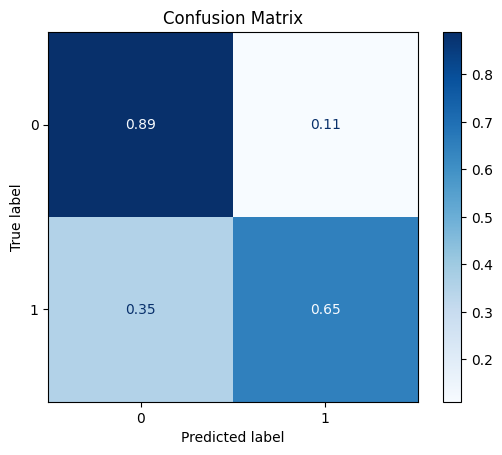
ROC AUC: 0.7821296607322807
Precision: 0.6132132849047058
Recall: 0.7676184077930803
F1: 0.6451779388650385
Accuracy: 0.8760330578512396
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
DataNormalizer
Next we try DataNormalizer, which will scale the vectors to have unit norm (i.e. vector of length one). This is often used in text classification, but we will use it here as well. But it does not improve the results.
[24]:
scaler = dp.DataNormalizer()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
No columns specified for imputation. These columns have been automatically identified:
[]
TEST SET:
[[1089 56]
[ 6 59]]
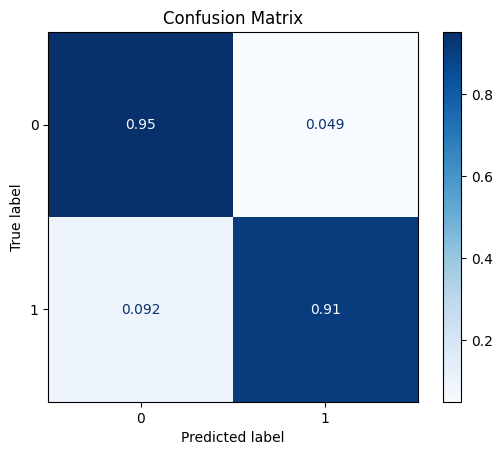
ROC AUC: 0.9453745381256298
Precision: 0.7537820131030375
Recall: 0.9293920053745381
F1: 0.8139384920634921
Accuracy: 0.9487603305785124
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
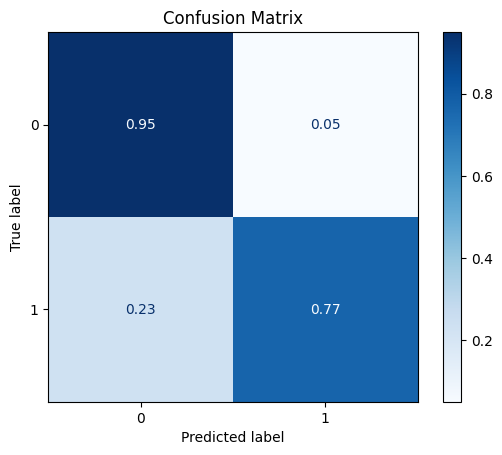
DataStandardScaler
The standard scaler sets the mean to zero and scales the vectors to have unit variance.
[25]:
scaler = dp.DataStandardScaler()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1088 57]
[ 15 50]]
ROC AUC: 0.8714746388982196
Precision: 0.7268452224604096
Recall: 0.8597245549210615
F1: 0.7746834395431599
Accuracy: 0.9404958677685951
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
DataQuantileTransformer
The Quantile Transformer transforms the each feature to have a normal distribution.
[26]:
scaler = dp.DataQuantileTransformer()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1113 32]
[ 10 55]]
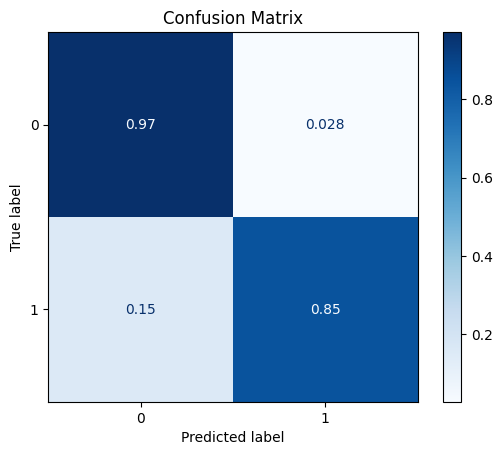
ROC AUC: 0.917991266375546
Precision: 0.8116395942723207
Recall: 0.9091031239502856
F1: 0.8525828460038987
Accuracy: 0.9652892561983472
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.6
Threshold used: 0.2
DataPowerTransformer
The power transformer makes the data more Gaussian-like (by default using the Yeo-Johnson transform).
[27]:
scaler = dp.DataPowerTransformer()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1118 27]
[ 7 58]]
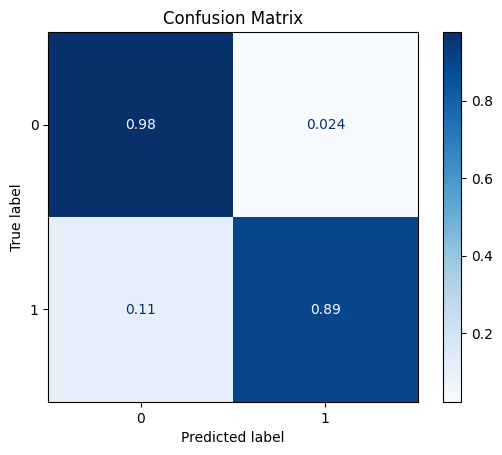
ROC AUC: 0.9416392341283171
Precision: 0.8380653594771241
Recall: 0.9343634531407458
F1: 0.8791776798825257
Accuracy: 0.971900826446281
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
DataRobustScaler
The robust scaler centers the data (median=zero) and scales the data based on the interquartile range (IQR).
[28]:
scaler = dp.DataRobustScaler()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1121 24]
[ 5 60]]
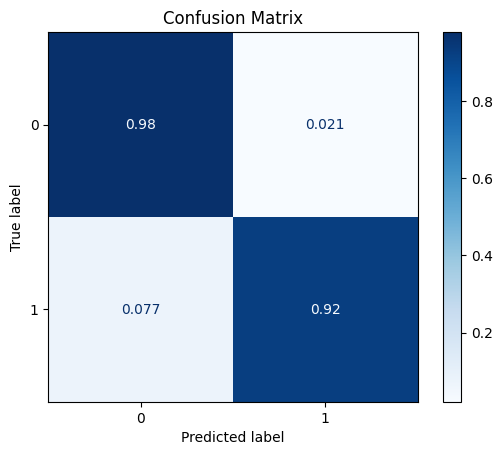
ROC AUC: 0.9579711118575748
Precision: 0.8549226084750063
Recall: 0.9510581121934834
F1: 0.8962997112704985
Accuracy: 0.9760330578512396
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
As we can see, this transformation had some impact in the results (depends on the seed used) when we use KNN. Let’s check how this data transformation impacts the XGBoost model:
[29]:
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="xgb", train_result=False, plot_pr=False)
TEST SET:
[[1115 30]
[ 6 59]]
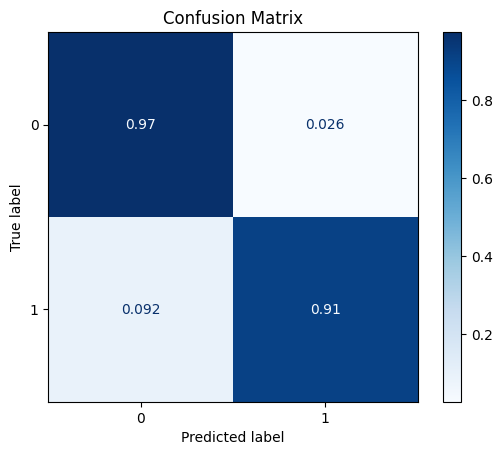
ROC AUC: 0.9681558616056434
Precision: 0.8287844921769287
Recall: 0.9407457171649312
F1: 0.8751733703190014
Accuracy: 0.9702479338842975
Optimal Threshold (ROC curve): 0.15434882044792175
Optimal Threshold (Precision x Recall curve): 0.1898476928472519
Threshold used: 0.15434882044792175
As depicted above, the results for the XGBoost remains the same as before. This shows us that data transformations are more impactful in certain models and less meaningful in others.
4 - Feature Selection
By using data scaling (Robust Scaler), we managed to get a small performance increase. We now proceed to use feature selection over the dataset and see if we can manage to get an even higher performance. We start out with 5 features, and use feature selection to remove unneeded features.
[30]:
feat_sel = dp.CatBoostSelection(steps=5, verbose=False)
feat_sel.fit(X=train_x_scl, y=train_y)
train_x_sel = feat_sel.transform(train_x_scl)
test_x_sel = feat_sel.transform(test_x_scl)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/catboost/core.py:1222: FutureWarning: iteritems is deprecated and will be removed in a future version. Use .items instead.
self._init_pool(data, label, cat_features, text_features, embedding_features, pairs, weight,
The number of features selection steps (5) is greater than the number of features to eliminate (3). The number of steps was reduced to 3.
[31]:
feat_sel.get_selected_features()
[31]:
['Mean_Green', 'Mean_Red', 'Mean_NIR', 'SD_pan']
Here, CatBoostSelection feature selection has removed one of the features (GLCM_pan).
[32]:
model = train_model_plot_results(train_x_sel, train_y, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1124 21]
[ 5 60]]
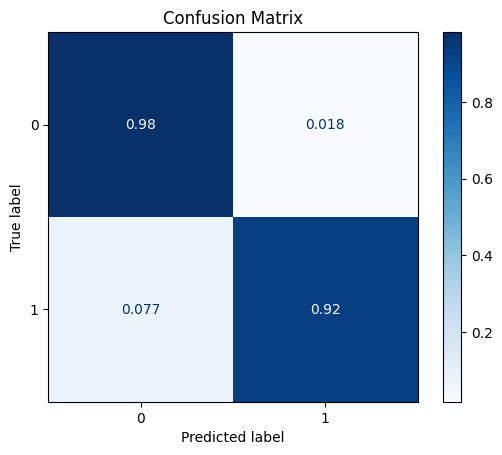
ROC AUC: 0.9586362109506215
Precision: 0.8681560213889709
Recall: 0.9523681558616057
F1: 0.9052421055168011
Accuracy: 0.9785123966942149
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
From the previous results, we can see that we removed one of the features, but without any noticeable gains in performance. However, we are now achieving the same metrics using less data, so we can already call it successful pre-processing step.
5 - Synthetic Data
imblearn Library
With unbalanced classes, we can instead choose to create synthetic data for the minority class. Here we use Rebalance, which connects to the imblearn library.
[33]:
train_y.value_counts()
[33]:
0 3433
1 196
Name: class, dtype: int64
[34]:
rebalance = dp.Rebalance(
X=train_x_sel,
y=train_y,
strategy_over={0:3433, 1:400},
over_sampler=True,
under_sampler=False
)
train_x_res, train_y_res = rebalance.fit_resample()
train_y_res.value_counts()
No columns specified for imputation. These columns have been automatically identified:
[]
Running oversampling...
...finished
[34]:
0 3433
1 400
Name: class, dtype: int64
[35]:
model = train_model_plot_results(train_x_res, train_y_res, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1122 23]
[ 5 60]]
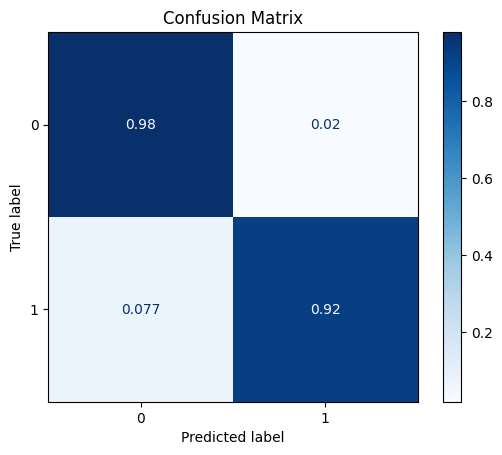
ROC AUC: 0.9583338931810548
Precision: 0.8592275045167359
Recall: 0.9514947934161908
F1: 0.8992434335744195
Accuracy: 0.9768595041322314
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
Similarly to the feature selection step, using the Rebalance class didn’t provide any considerable gains.
Creating Artificial Data using Deep Learning
CTGAN
Let’s now test rebalancing the dataset using CTGAN:
[36]:
synth = dp.Synthesizer(
X=train_x_sel,
y=train_y,
epochs=200,
model="ctgan",
load_existing=False
)
conditions = {label_col:1} # create more of the undersampled class
syn_train_x, syn_train_y = synth.fit_resample(X=train_x_sel, y=train_y, n_samples=200, conditions=conditions)
syn_train_y.value_counts()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
Sampling conditions: 0%| | 0/200 [00:00<?, ?it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:608: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for group, dataframe in grouped_conditions:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:639: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for transformed_group, transformed_dataframe in transformed_groups:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 100%|██████████| 200/200 [00:00<00:00, 1615.28it/s]
[36]:
0 3433
1 396
Name: class, dtype: int64
[37]:
model = train_model_plot_results(syn_train_x, syn_train_y, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1127 18]
[ 9 56]]
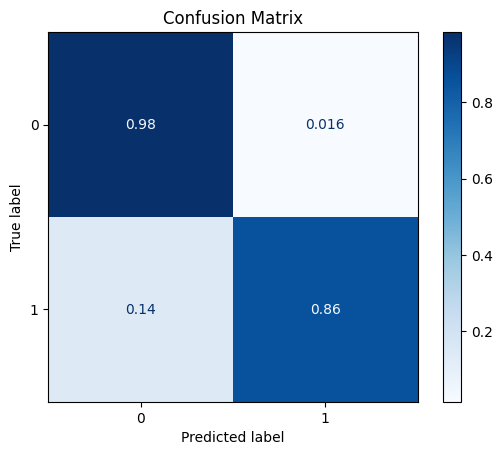
ROC AUC: 0.9528787369835405
Precision: 0.8744171107727445
Recall: 0.9229089687604972
F1: 0.8969592410245412
Accuracy: 0.9776859504132231
Optimal Threshold (ROC curve): 0.4
Optimal Threshold (Precision x Recall curve): 0.6
Threshold used: 0.4
Let’s try creating more artificial instances and check how this impacts the model’s performance.
[38]:
conditions = {label_col:1} # create more of the undersampled class
syn_train_x, syn_train_y = synth.fit_resample(X=train_x_sel, y=train_y, n_samples=2000, conditions=conditions)
model = train_model_plot_results(syn_train_x, syn_train_y, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
Sampling conditions: 0%| | 0/2000 [00:00<?, ?it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:608: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for group, dataframe in grouped_conditions:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:639: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for transformed_group, transformed_dataframe in transformed_groups:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 64%|██████▍ | 1276/2000 [00:00<00:00, 11106.77it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 100%|██████████| 2000/2000 [00:00<00:00, 9766.39it/s]
TEST SET:
[[1082 63]
[ 4 61]]
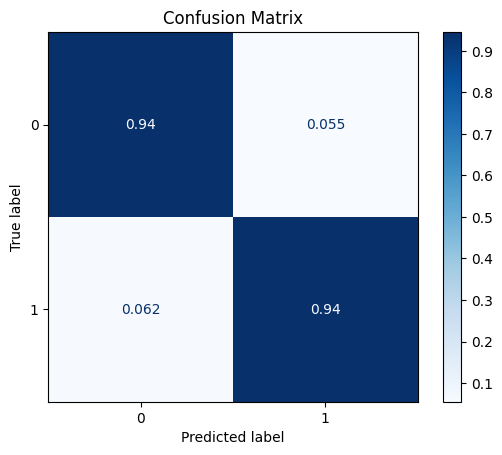
ROC AUC: 0.9773328854551563
Precision: 0.7441261213093329
Recall: 0.9417198522002015
F1: 0.8077356347190503
Accuracy: 0.9446280991735537
Optimal Threshold (ROC curve): 0.6
Optimal Threshold (Precision x Recall curve): 1.0
Threshold used: 0.6
TVAE
[39]:
synth = dp.Synthesizer(
X=train_x_sel,
y=train_y,
epochs=200,
model="tvae",
load_existing=False
)
conditions = {label_col:1} # create more of the undersampled class
syn2_train_x, syn2_train_y = synth.fit_resample(X=train_x_sel, y=train_y, n_samples=200, conditions=conditions)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
Sampling conditions: 0%| | 0/200 [00:00<?, ?it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:608: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for group, dataframe in grouped_conditions:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:639: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for transformed_group, transformed_dataframe in transformed_groups:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 46%|████▋ | 93/200 [00:00<00:00, 781.54it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 98%|█████████▊| 196/200 [00:00<00:00, 892.97it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 100%|██████████| 200/200 [00:00<00:00, 820.42it/s]
[40]:
model = train_model_plot_results(syn2_train_x, syn2_train_y, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[1067 78]
[ 3 62]]
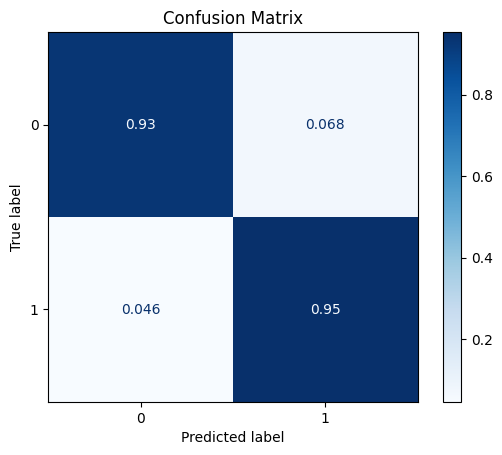
ROC AUC: 0.9687000335908633
Precision: 0.7200267022696929
Recall: 0.942861941551898
F1: 0.7841546000110114
Accuracy: 0.9330578512396694
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.8
Threshold used: 0.2