[1]:
import pandas as pd
import numpy as np
import uci_dataset as database
import torch
from raimitigations.utils import train_model_plot_results, split_data
import raimitigations.dataprocessing as dp
Case Study 3
Fixing a seed
To avoid randomness in the following experiments, we’ll fix the seeds to guarantee that the results obtained are the same each time we run this notebook. Feel free to comment the next cell or test different seeds to see how this affects the results.
[2]:
import random
SEED = 45
torch.manual_seed(SEED)
np.random.seed(SEED)
random.seed(SEED)
1 - Understanding the Data
Case study 3 examines the thyroid disease dataset. We will explore how this dataset with the dataprocessing library.
[3]:
df = database.load_thyroid_disease()
label_col = "sick-euthyroid"
df
[3]:
| sick-euthyroid | age | sex | on_thyroxine | query_on_thyroxine | on_antithyroid_medication | thyroid_surgery | query_hypothyroid | query_hyperthyroid | pregnant | ... | T3_measured | T3 | TT4_measured | TT4 | T4U_measured | T4U | FTI_measured | FTI | TBG_measured | TBG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | sick-euthyroid | 72.0 | M | f | f | f | f | f | f | f | ... | y | 1.0 | y | 83.0 | y | 0.95 | y | 87.0 | n | NaN |
| 1 | sick-euthyroid | 45.0 | F | f | f | f | f | f | f | f | ... | y | 1.0 | y | 82.0 | y | 0.73 | y | 112.0 | n | NaN |
| 2 | sick-euthyroid | 64.0 | F | f | f | f | f | f | f | f | ... | y | 1.0 | y | 101.0 | y | 0.82 | y | 123.0 | n | NaN |
| 3 | sick-euthyroid | 56.0 | M | f | f | f | f | f | f | f | ... | y | 0.8 | y | 76.0 | y | 0.77 | y | 99.0 | n | NaN |
| 4 | sick-euthyroid | 78.0 | F | t | f | f | f | t | f | f | ... | y | 0.3 | y | 87.0 | y | 0.95 | y | 91.0 | n | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 3158 | negative | 40.0 | F | f | f | f | f | f | f | f | ... | y | 1.2 | y | 76.0 | y | 0.90 | y | 84.0 | n | NaN |
| 3159 | negative | 69.0 | F | f | f | f | f | f | f | f | ... | y | 1.8 | y | 126.0 | y | 1.02 | y | 124.0 | n | NaN |
| 3160 | negative | 58.0 | F | f | f | f | f | f | f | f | ... | y | 1.7 | y | 86.0 | y | 0.91 | y | 95.0 | n | NaN |
| 3161 | negative | 29.0 | F | f | f | f | f | f | f | f | ... | y | 1.8 | y | 99.0 | y | 1.01 | y | 98.0 | n | NaN |
| 3162 | negative | 56.0 | F | t | f | f | f | f | f | f | ... | y | 1.8 | y | 139.0 | y | 0.97 | y | 143.0 | n | NaN |
3163 rows × 26 columns
[4]:
df[label_col] = df[label_col].replace({"sick-euthyroid": 1, "negative": 0})
[5]:
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 3163 entries, 0 to 3162
Data columns (total 26 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 sick-euthyroid 3163 non-null int64
1 age 2717 non-null float64
2 sex 3090 non-null object
3 on_thyroxine 3163 non-null object
4 query_on_thyroxine 3163 non-null object
5 on_antithyroid_medication 3163 non-null object
6 thyroid_surgery 3163 non-null object
7 query_hypothyroid 3163 non-null object
8 query_hyperthyroid 3163 non-null object
9 pregnant 3163 non-null object
10 sick 3163 non-null object
11 tumor 3163 non-null object
12 lithium 3163 non-null object
13 goitre 3163 non-null object
14 TSH_measured 3163 non-null object
15 TSH 2695 non-null float64
16 T3_measured 3163 non-null object
17 T3 2468 non-null float64
18 TT4_measured 3163 non-null object
19 TT4 2914 non-null float64
20 T4U_measured 3163 non-null object
21 T4U 2915 non-null float64
22 FTI_measured 3163 non-null object
23 FTI 2916 non-null float64
24 TBG_measured 3163 non-null object
25 TBG 260 non-null float64
dtypes: float64(7), int64(1), object(18)
memory usage: 642.6+ KB
[6]:
df['query_on_thyroxine'].value_counts()
[6]:
f 3108
t 55
Name: query_on_thyroxine, dtype: int64
The dataset is imbalanced, as most patients in the dataset do not have thyroid disease.
[7]:
counts = df['query_on_thyroxine'].value_counts().values
counts
[7]:
array([3108, 55])
[8]:
cor_feat = dp.CorrelatedFeatures(
method_num_num=["spearman", "pearson", "kendall"], # Used for Numerical x Numerical correlations
num_corr_th=0.9, # Used for Numerical x Numerical correlations
num_pvalue_th=0.05, # Used for Numerical x Numerical correlations
method_num_cat="model", # Used for Numerical x Categorical correlations
model_metrics=["f1", "auc"], # Used for Numerical x Categorical correlations
metric_th=0.9, # Used for Numerical x Categorical correlations
cat_corr_th=0.9, # Used for Categorical x Categorical correlations
cat_pvalue_th=0.01, # Used for Categorical x Categorical correlations
json_summary="./corr_json/c3_summary.json",
json_corr="./corr_json/c3_corr.json",
json_uncorr="./corr_json/c3_uncorr.json"
)
cor_feat.fit(df=df, label_col=label_col)
[8]:
<raimitigations.dataprocessing.feat_selection.correlated_features.CorrelatedFeatures at 0x7fcaa4b54880>
Remember to look through the JSON files generated in the previous cell. Correlations have been found, which can be seen in c3_corr.json.
2 - Basic Pre-Processing
Encode Categorical Variables
The dataset contains a number of columns that are categorical. EncoderOHE is used to perform One Hot Encoding on the dataset.
[9]:
# Encode the categorical columns using One-Hot Encoding
enc_ohe = dp.EncoderOHE()
enc_ohe.fit(df)
proc_df = enc_ohe.transform(df)
proc_df
No columns specified for encoding. These columns have been automatically identfied as the following:
['sex', 'on_thyroxine', 'query_on_thyroxine', 'on_antithyroid_medication', 'thyroid_surgery', 'query_hypothyroid', 'query_hyperthyroid', 'pregnant', 'sick', 'tumor', 'lithium', 'goitre', 'TSH_measured', 'T3_measured', 'TT4_measured', 'T4U_measured', 'FTI_measured', 'TBG_measured']
[9]:
| sick-euthyroid | age | TSH | T3 | TT4 | T4U | FTI | TBG | sex_M | sex_nan | ... | sick_t | tumor_t | lithium_t | goitre_t | TSH_measured_y | T3_measured_y | TT4_measured_y | T4U_measured_y | FTI_measured_y | TBG_measured_y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 72.0 | NaN | 1.0 | 83.0 | 0.95 | 87.0 | NaN | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 |
| 1 | 1 | 45.0 | 1.90 | 1.0 | 82.0 | 0.73 | 112.0 | NaN | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| 2 | 1 | 64.0 | 0.09 | 1.0 | 101.0 | 0.82 | 123.0 | NaN | 0 | 0 | ... | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| 3 | 1 | 56.0 | 0.00 | 0.8 | 76.0 | 0.77 | 99.0 | NaN | 1 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| 4 | 1 | 78.0 | 2.60 | 0.3 | 87.0 | 0.95 | 91.0 | NaN | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 3158 | 0 | 40.0 | 2.10 | 1.2 | 76.0 | 0.90 | 84.0 | NaN | 0 | 0 | ... | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| 3159 | 0 | 69.0 | 2.60 | 1.8 | 126.0 | 1.02 | 124.0 | NaN | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| 3160 | 0 | 58.0 | 5.80 | 1.7 | 86.0 | 0.91 | 95.0 | NaN | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| 3161 | 0 | 29.0 | 0.80 | 1.8 | 99.0 | 1.01 | 98.0 | NaN | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| 3162 | 0 | 56.0 | 0.00 | 1.8 | 139.0 | 0.97 | 143.0 | NaN | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
3163 rows × 27 columns
Impute Missing Data
A number of features contain missing values. We will use BasicImputer to fill these values with -1
[10]:
proc_df.isna().sum()
[10]:
sick-euthyroid 0
age 446
TSH 468
T3 695
TT4 249
T4U 248
FTI 247
TBG 2903
sex_M 0
sex_nan 0
on_thyroxine_t 0
query_on_thyroxine_t 0
on_antithyroid_medication_t 0
thyroid_surgery_t 0
query_hypothyroid_t 0
query_hyperthyroid_t 0
pregnant_t 0
sick_t 0
tumor_t 0
lithium_t 0
goitre_t 0
TSH_measured_y 0
T3_measured_y 0
TT4_measured_y 0
T4U_measured_y 0
FTI_measured_y 0
TBG_measured_y 0
dtype: int64
[11]:
imputer = dp.BasicImputer(numerical={'missing_values':np.nan,
'strategy':'constant',
'fill_value':-1})
imputer.fit(proc_df)
proc_df = imputer.transform(proc_df)
No columns specified for imputation. These columns have been automatically identified:
['age', 'TSH', 'T3', 'TT4', 'T4U', 'FTI', 'TBG']
Split Dataset
[12]:
train_x, test_x, train_y, test_y = split_data(proc_df, label_col, test_size=0.25)
2 - Baseline Models
In this example, we have an imbalanced dataset (most patients do not have thyroid disease). While we will take a look at a number of different metrics, we will be focused on improved the F1 score for this dataset.
After splitting the data into train and test sets, we will build two baseline models, one with XGBoost, and the other with KNN.
[13]:
model = train_model_plot_results(train_x, train_y, test_x, test_y, model="xgb", train_result=False, plot_pr=False)
TEST SET:
[[704 14]
[ 9 64]]
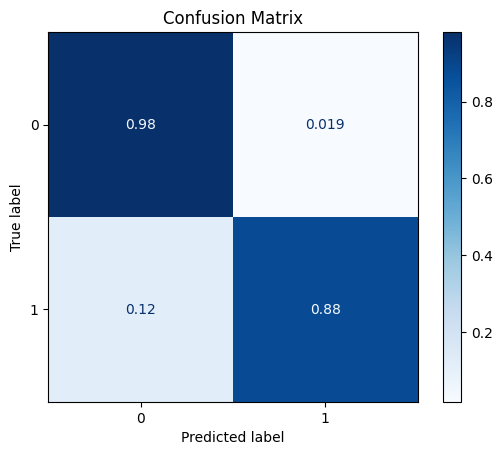
ROC AUC: 0.9617182432174611
Precision: 0.9039450498076024
Recall: 0.9286068607623916
F1: 0.9158047213776316
Accuracy: 0.9709228824273072
Optimal Threshold (ROC curve): 0.29627740383148193
Optimal Threshold (Precision x Recall curve): 0.37958627939224243
Threshold used: 0.29627740383148193
[14]:
model = train_model_plot_results(train_x, train_y, test_x, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[536 182]
[ 27 46]]
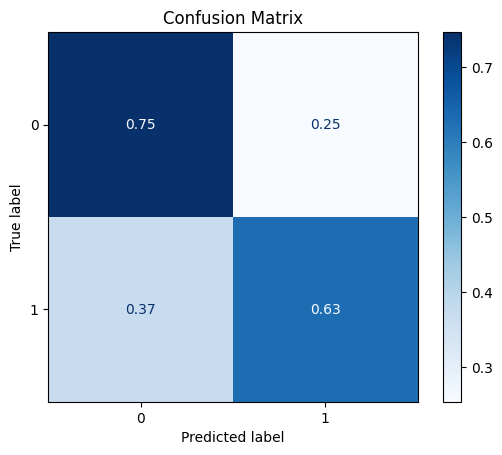
ROC AUC: 0.7110218643873774
Precision: 0.5768985073696675
Recall: 0.6883275460754761
F1: 0.5712470272134779
Accuracy: 0.7357774968394437
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
3 - Data Transformation
We will perform a number of different feature transformations, using the Scalers from dataprocessing.
DataMinMaxScaler
The first data transformation we will perform is a MinMaxScaler, which will scale each feature to have a range between zero and one
[15]:
scaler = dp.DataMinMaxScaler()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[611 107]
[ 13 60]]
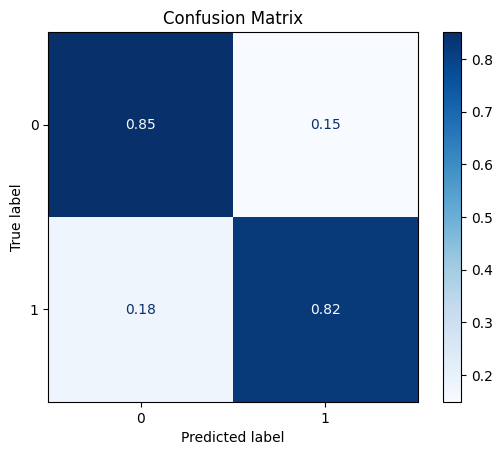
ROC AUC: 0.8665528293967262
Precision: 0.6692240518962076
Recall: 0.8364463692906475
F1: 0.7052906110283159
Accuracy: 0.8482932996207333
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
DataNormalizer
Next we try DataNormalizer, which will scale the vectors to have unit norm (i.e. vector of length one). This is often used in text classification, but we will use it here as well.
[16]:
scaler = dp.DataNormalizer()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
No columns specified for imputation. These columns have been automatically identified:
[]
TEST SET:
[[541 177]
[ 26 47]]
/home/matheus/Dropbox/Matheus/Microsoft_Research/Code/ResponsibleAI/ResponsibleAI/git/responsible-ai-toolbox-mitigations/raimitigations/utils/metric_utils.py:185: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
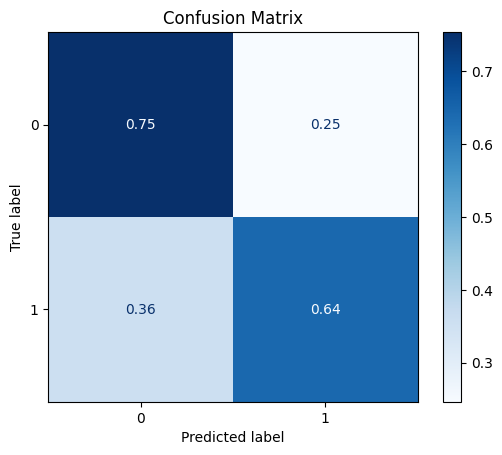
ROC AUC: 0.7177090090433853
Precision: 0.581983024691358
Recall: 0.698658755294387
F1: 0.5792608314009092
Accuracy: 0.7433628318584071
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 1.0
Threshold used: 0.2
DataQuantileTransformer
The Quantile Transformer transforms the each feature to have a normal distribution.
[17]:
scaler = dp.DataQuantileTransformer()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[598 120]
[ 9 64]]
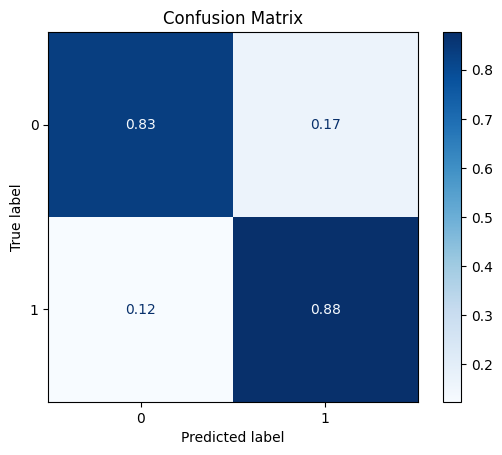
ROC AUC: 0.8824836875643911
Precision: 0.6664995344173054
Recall: 0.8547907047735337
F1: 0.7003479920710668
Accuracy: 0.8369152970922883
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
DataRobustScaler
The robust scaler centers the data (median=zero) and scales the data based on the interquartile range (IQR).
[18]:
scaler = dp.DataRobustScaler()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[611 107]
[ 9 64]]
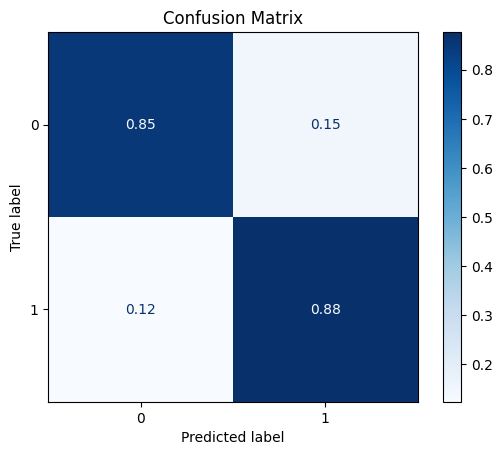
ROC AUC: 0.8919468081047048
Precision: 0.6798764384078475
Recall: 0.8638436295646201
F1: 0.7189468009507707
Accuracy: 0.8533501896333755
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
DataPowerTransformer
The power transformer makes the data more Gaussian-like (by default using the Yeo-Johnson transform).
[19]:
scaler = dp.DataPowerTransformer()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[590 128]
[ 11 62]]
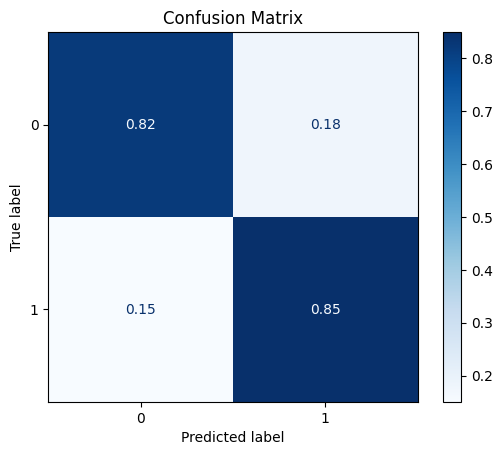
ROC AUC: 0.8798221849124279
Precision: 0.654006480427358
Recall: 0.8355210439958789
F1: 0.6830500119632053
Accuracy: 0.8242730720606827
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
DataStandardScaler
The standard scaler sets the mean to zero and scales the vectors to have unit variance.
[20]:
scaler = dp.DataStandardScaler()
scaler.fit(train_x)
train_x_scl = scaler.transform(train_x)
test_x_scl = scaler.transform(test_x)
model = train_model_plot_results(train_x_scl, train_y, test_x_scl, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[605 113]
[ 8 65]]
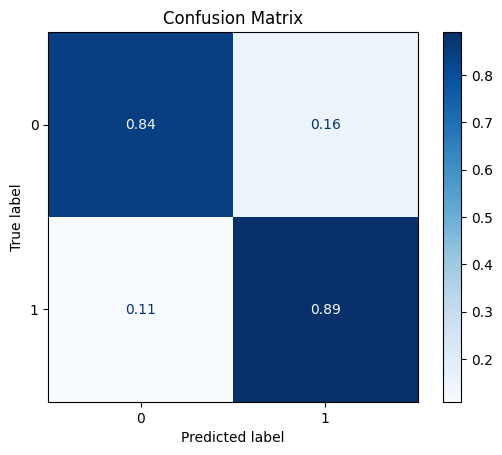
ROC AUC: 0.9002842751936504
Precision: 0.6760589841816815
Recall: 0.866514671652612
F1: 0.7135095979717494
Accuracy: 0.8470290771175727
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
From the previous results, we can see that the DataStandardScaler managed to get a considerable improvement over the baseline model. We’ll use this scaled dataset in the following experiments.
4 - Feature Selection
Next, we perform a feature selection, which removes features from the dataset.
[21]:
feat_sel = dp.CatBoostSelection(steps=5, verbose=False)
feat_sel.fit(X=train_x_scl, y=train_y)
train_x_sel = feat_sel.transform(train_x_scl)
test_x_sel = feat_sel.transform(test_x_scl)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/catboost/core.py:1222: FutureWarning: iteritems is deprecated and will be removed in a future version. Use .items instead.
self._init_pool(data, label, cat_features, text_features, embedding_features, pairs, weight,
[22]:
feat_sel.get_selected_features()
[22]:
['age',
'TSH',
'T3',
'TT4',
'FTI',
'TBG',
'sex_nan',
'on_thyroxine_t',
'on_antithyroid_medication_t',
'thyroid_surgery_t',
'query_hypothyroid_t',
'query_hyperthyroid_t',
'pregnant_t',
'sick_t',
'lithium_t',
'goitre_t',
'T3_measured_y']
We can see that we managed to remove a considerable amount of features from the dataset. Now, let’s see if we manage to get any improvement in the metrics.
[23]:
model = train_model_plot_results(train_x_sel, train_y, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[626 92]
[ 9 64]]
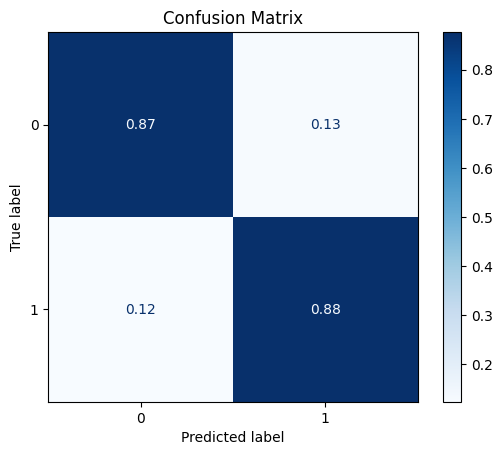
ROC AUC: 0.907124050826115
Precision: 0.6980415909549768
Recall: 0.8742893120158737
F1: 0.7421515183790186
Accuracy: 0.8723135271807838
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
We can see that the CatBoostSelection class managed to improve even further the results obtained with the scaled data, where we have a higher F1 score and a similar ROC AUC. Note that we are now using a lot less data, since we removed several features from the dataset.
5 - Synthetic Data
Since we have an imbalanced dataset, we can generate instances of the minority class to improve the balance between the classes. Below, we increase the quantity of minority instances (patients with disease) from 220 to 400.
imblearn Library
[24]:
train_y.value_counts()
[24]:
0 2152
1 220
Name: sick-euthyroid, dtype: int64
[25]:
rebalance = dp.Rebalance(
X=train_x_sel,
y=train_y,
strategy_over={0:2152, 1:400},
over_sampler=True,
under_sampler=False
)
train_x_res, train_y_res = rebalance.fit_resample()
train_y_res.value_counts()
No columns specified for imputation. These columns have been automatically identified:
[]
Running oversampling...
...finished
[25]:
0 2152
1 400
Name: sick-euthyroid, dtype: int64
[26]:
model = train_model_plot_results(train_x_res, train_y_res, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[623 95]
[ 8 65]]
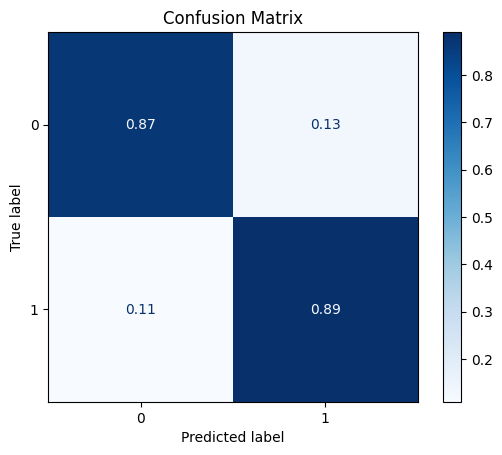
ROC AUC: 0.9110542984698745
Precision: 0.696785855784469
Recall: 0.8790494905941161
F1: 0.7407935300985947
Accuracy: 0.8697850821744627
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
Notice that the Rebalance class managed to slightly improve the ROC AUC compared to the feature selection step, while maintaining a similar F1 score.
Creating Artificial Data using Deep Learning
CTGAN
[27]:
synth = dp.Synthesizer(
X=train_x_sel,
y=train_y,
epochs=200,
model="ctgan",
load_existing=False
)
conditions = {label_col:1} # create more of the undersampled class
syn_train_x, syn_train_y = synth.fit_resample(X=train_x_sel, y=train_y, n_samples=200, conditions=conditions)
syn_train_y.value_counts()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
Sampling conditions: 0%| | 0/200 [00:00<?, ?it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:608: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for group, dataframe in grouped_conditions:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:639: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for transformed_group, transformed_dataframe in transformed_groups:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 100%|██████████| 200/200 [00:00<00:00, 1783.00it/s]
[27]:
0 2152
1 420
Name: sick-euthyroid, dtype: int64
[28]:
model = train_model_plot_results(syn_train_x, syn_train_y, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[580 138]
[ 7 66]]
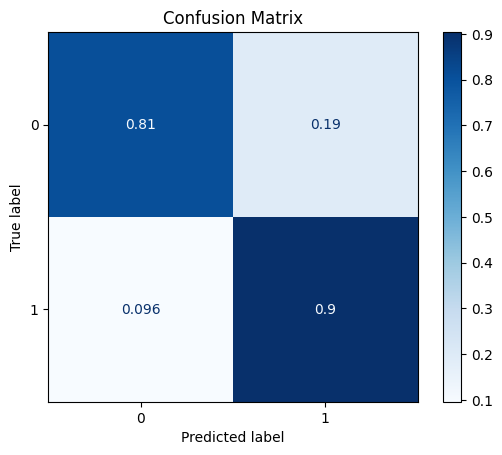
ROC AUC: 0.9070572747739154
Precision: 0.6558021845876341
Recall: 0.8559545159690158
F1: 0.6827115924588849
Accuracy: 0.8166877370417194
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2
[29]:
conditions = {label_col:1} # create more of the undersampled class
syn_train_x, syn_train_y = synth.fit_resample(X=train_x_sel, y=train_y, n_samples=600, conditions=conditions)
model = train_model_plot_results(syn_train_x, syn_train_y, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
Sampling conditions: 0%| | 0/600 [00:00<?, ?it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:608: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for group, dataframe in grouped_conditions:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:639: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for transformed_group, transformed_dataframe in transformed_groups:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 100%|██████████| 600/600 [00:00<00:00, 4369.32it/s]
TEST SET:
[[537 181]
[ 5 68]]
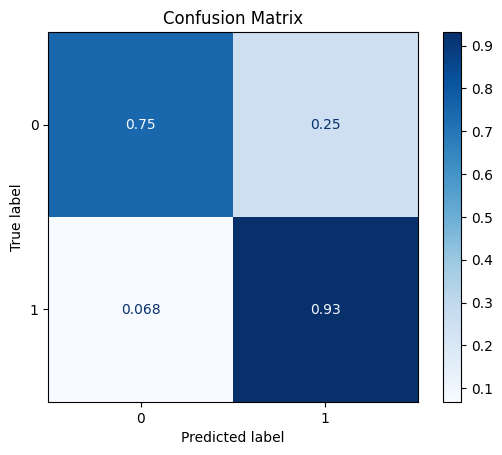
ROC AUC: 0.9090414774678521
Precision: 0.6319336386134946
Recall: 0.8397088564124089
F1: 0.6373706004140787
Accuracy: 0.7648546144121365
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.6
Threshold used: 0.2
TVAE
Finally, we generate artificial instances using TVAE (a variational auto encoder for generating tabular data).
[30]:
synth = dp.Synthesizer(
X=train_x_sel,
y=train_y,
epochs=200,
model="tvae",
load_existing=False
)
conditions = {label_col:1} # create more of the undersampled class
syn2_train_x, syn2_train_y = synth.fit_resample(X=train_x_sel, y=train_y, n_samples=200, conditions=conditions)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:286: ConvergenceWarning: Initialization 1 did not converge. Try different init parameters, or increase max_iter, tol or check for degenerate data.
warnings.warn(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sklearn/mixture/_base.py:131: ConvergenceWarning: Number of distinct clusters (2) found smaller than n_clusters (10). Possibly due to duplicate points in X.
cluster.KMeans(
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:111: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
data[column_name] = data[column_name].to_numpy().flatten()
Sampling conditions: 0%| | 0/200 [00:00<?, ?it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:608: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for group, dataframe in grouped_conditions:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/sdv/tabular/base.py:639: FutureWarning: In a future version of pandas, a length 1 tuple will be returned when iterating over a groupby with a grouper equal to a list of length 1. Don't supply a list with a single grouper to avoid this warning.
for transformed_group, transformed_dataframe in transformed_groups:
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 46%|████▌ | 91/200 [00:00<00:00, 758.20it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 99%|█████████▉| 198/200 [00:00<00:00, 810.50it/s]/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
/home/matheus/miniconda3/envs/rai/lib/python3.9/site-packages/ctgan/data_transformer.py:149: FutureWarning: In a future version, `df.iloc[:, i] = newvals` will attempt to set the values inplace instead of always setting a new array. To retain the old behavior, use either `df[df.columns[i]] = newvals` or, if columns are non-unique, `df.isetitem(i, newvals)`
data.iloc[:, 1] = np.argmax(column_data[:, 1:], axis=1)
Sampling conditions: 100%|██████████| 200/200 [00:00<00:00, 667.71it/s]
[31]:
model = train_model_plot_results(syn2_train_x, syn2_train_y, test_x_sel, test_y, model="knn", train_result=False, plot_pr=False)
TEST SET:
[[606 112]
[ 9 64]]
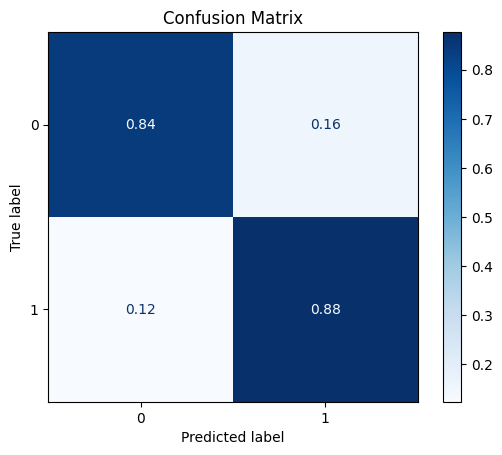
ROC AUC: 0.9024497271721296
Precision: 0.6745011086474502
Recall: 0.8603617354142024
F1: 0.7116417658631524
Accuracy: 0.8470290771175727
Optimal Threshold (ROC curve): 0.2
Optimal Threshold (Precision x Recall curve): 0.4
Threshold used: 0.2