Case Study 2 - Multiple Runs
This notebook uses the seaborn, which is not listed as a requirement for this library. Therefore, make sure to install it before running this notebook. To install it, use the following:
> pip install seaborn
This notebook, as in case2.ipynb, we will explore the wilt dataset. Similar to case1.ipynb/case1_stat.ipynb, we will run the same experiment multiple times in order to see if a preprocessing step improves the model’s performance with statistical significance.
0 - Defining Basic Functionalities
[1]:
import pandas as pd
import numpy as np
import uci_dataset as database
from matplotlib.pyplot import figure
from raimitigations.utils import train_model_fetch_results, split_data
import raimitigations.dataprocessing as dp
import case_study_utils as cs
N_EXEC = 50
USE_AUC_TH = False
[2]:
def test_base(df, label_col, n_exec, model_name):
result_list = []
for n in range(n_exec):
train_x, test_x, train_y, test_y = split_data(df, label_col, test_size=0.25)
result = train_model_fetch_results(train_x, train_y, test_x, test_y, model_name, USE_AUC_TH)
result_list.append(result)
result_stat = cs.result_statistics(result_list)
return result_stat
# -----------------------------------
def test_corr(df, label_col, n_exec, model_name):
result_list = []
proc_df = cs.remove_corr_feat(df, label_col)
for n in range(n_exec):
train_x, test_x, train_y, test_y = split_data(proc_df, label_col, test_size=0.25)
result = train_model_fetch_results(train_x, train_y, test_x, test_y, model_name, USE_AUC_TH)
result_list.append(result)
result_stat = cs.result_statistics(result_list)
return result_stat
# -----------------------------------
def test_corr_transf(df, label_col, n_exec, scaler_ref, model_name, num_col=None):
result_list = []
proc_df = cs.remove_corr_feat(df, label_col)
for n in range(n_exec):
train_x, test_x, train_y, test_y = split_data(proc_df, label_col, test_size=0.25)
train_x, test_x = cs.transform_num_data(train_x, test_x, scaler_ref, num_col)
result = train_model_fetch_results(train_x, train_y, test_x, test_y, model_name, USE_AUC_TH)
result_list.append(result)
result_stat = cs.result_statistics(result_list)
return result_stat
# -----------------------------------
def test_smote_transf(df, label_col, n_exec, model_name, rcorr=True, scaler_ref=None, num_col=None, feat_sel_type=None, art_str=None, under=False):
result_list = []
proc_df = df
if rcorr:
proc_df = cs.remove_corr_feat(proc_df, label_col)
for n in range(n_exec):
train_x, test_x, train_y, test_y = split_data(proc_df, label_col, test_size=0.25)
if art_str is not None:
train_x, train_y = cs.artificial_smote(train_x, train_y, art_str, under)
if feat_sel_type is not None:
train_x, test_x = cs.feature_selection(train_x, train_y, test_x, feat_sel_type)
if scaler_ref is not None:
train_x, test_x = cs.transform_num_data(train_x, test_x, scaler_ref, num_col)
result = train_model_fetch_results(train_x, train_y, test_x, test_y, model_name, USE_AUC_TH)
result_list.append(result)
result_stat = cs.result_statistics(result_list)
return result_stat
# -----------------------------------
def test_ctgan_first(df, label_col, n_exec, model_name, rcorr=True, scaler_ref=None, num_col=None, feat_sel_type=None, art_str=None, savefile=None):
result_list = []
proc_df = df
if rcorr:
proc_df = cs.remove_corr_feat(proc_df, label_col)
for n in range(n_exec):
train_x, test_x, train_y, test_y = split_data(proc_df, label_col, test_size=0.25)
if art_str is not None:
train_x, train_y = cs.artificial_ctgan(train_x, train_y, art_str, savefile, epochs=1000)
if feat_sel_type is not None:
train_x, test_x = cs.feature_selection(train_x, train_y, test_x, feat_sel_type)
if scaler_ref is not None:
train_x, test_x = cs.transform_num_data(train_x, test_x, scaler_ref, num_col)
result = train_model_fetch_results(train_x, train_y, test_x, test_y, model_name, USE_AUC_TH)
result_list.append(result)
result_stat = cs.result_statistics(result_list)
return result_stat
1 - Understanding the Data
[3]:
df = database.load_wilt()
label_col = "class"
df[label_col] = df[label_col].replace({"w": 1, "n": 0})
df
[3]:
| class | GLCM_pan | Mean_Green | Mean_Red | Mean_NIR | SD_pan | |
|---|---|---|---|---|---|---|
| 0 | 1 | 120.362774 | 205.500000 | 119.395349 | 416.581395 | 20.676318 |
| 1 | 1 | 124.739583 | 202.800000 | 115.333333 | 354.333333 | 16.707151 |
| 2 | 1 | 134.691964 | 199.285714 | 116.857143 | 477.857143 | 22.496712 |
| 3 | 1 | 127.946309 | 178.368421 | 92.368421 | 278.473684 | 14.977453 |
| 4 | 1 | 135.431548 | 197.000000 | 112.690476 | 532.952381 | 17.604193 |
| ... | ... | ... | ... | ... | ... | ... |
| 495 | 0 | 123.554348 | 202.826087 | 106.391304 | 364.565217 | 17.314068 |
| 496 | 0 | 121.549028 | 276.220000 | 175.593333 | 402.620000 | 13.394574 |
| 497 | 0 | 119.076687 | 247.951220 | 113.365854 | 808.024390 | 24.830059 |
| 498 | 0 | 107.944444 | 197.000000 | 90.000000 | 451.000000 | 8.214887 |
| 499 | 0 | 119.731928 | 182.238095 | 74.285714 | 301.690476 | 22.944278 |
4839 rows × 6 columns
[ ]:
df.info()
[5]:
num_col = ["GLCM_pan", "Mean_Green", "Mean_Red", "Mean_NIR", "SD_pan"]
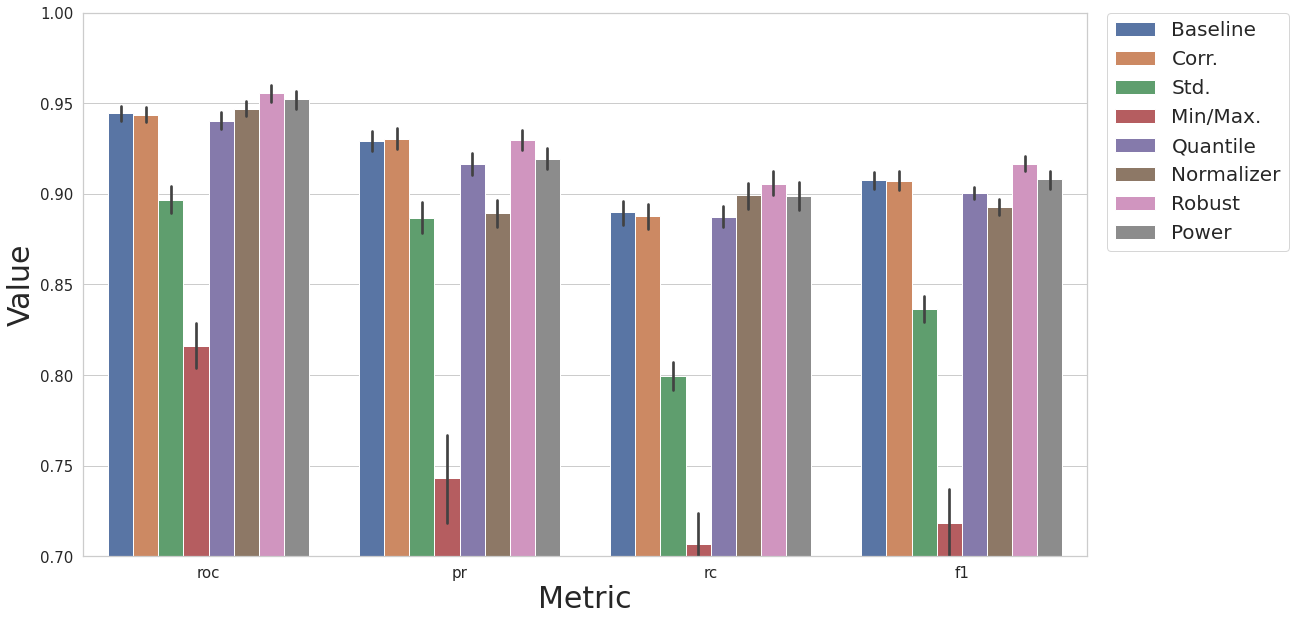
2 - Efficiency of Different Classes from raimitigations.dataprocessing lib
KNN Model
Data Transformations
Here we compare 8 different training pipelines with a KNN model. For each pipeline, 50 runs are performed, and the mean and stdev of the metrics are computed and plotted in the graph below.
baseline dataset
baseline dataset, removal of correlated features
baseline dataset, removal of correlated features, standard scaler
baseline dataset, removal of correlated features, minmax scaler
baseline dataset, removal of correlated features, quantile transformer
baseline dataset, removal of correlated features, data normalizer
baseline dataset, removal of correlated features, robust scaler
baseline dataset, removal of correlated features, power transformer
Note: Some of the experiments below take 10+ min to run. You may change N_EXEC to a smaller number to decrease the time it takes to run the notebook.
[6]:
MODEL_NAME = "knn"
N_EXEC = 50
result_base = test_base(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(None, result_base, "Baseline")
result_cor = test_corr(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(result_df, result_cor, "Corr.")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataStandardScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Std.")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataMinMaxScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Min/Max.")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataQuantileTransformer, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Quantile")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataNormalizer, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Normalizer")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataRobustScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Robust")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataPowerTransformer, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Power")
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
[8]:
cs.plot_results(result_df, y_lim=[0.7, 1.0])
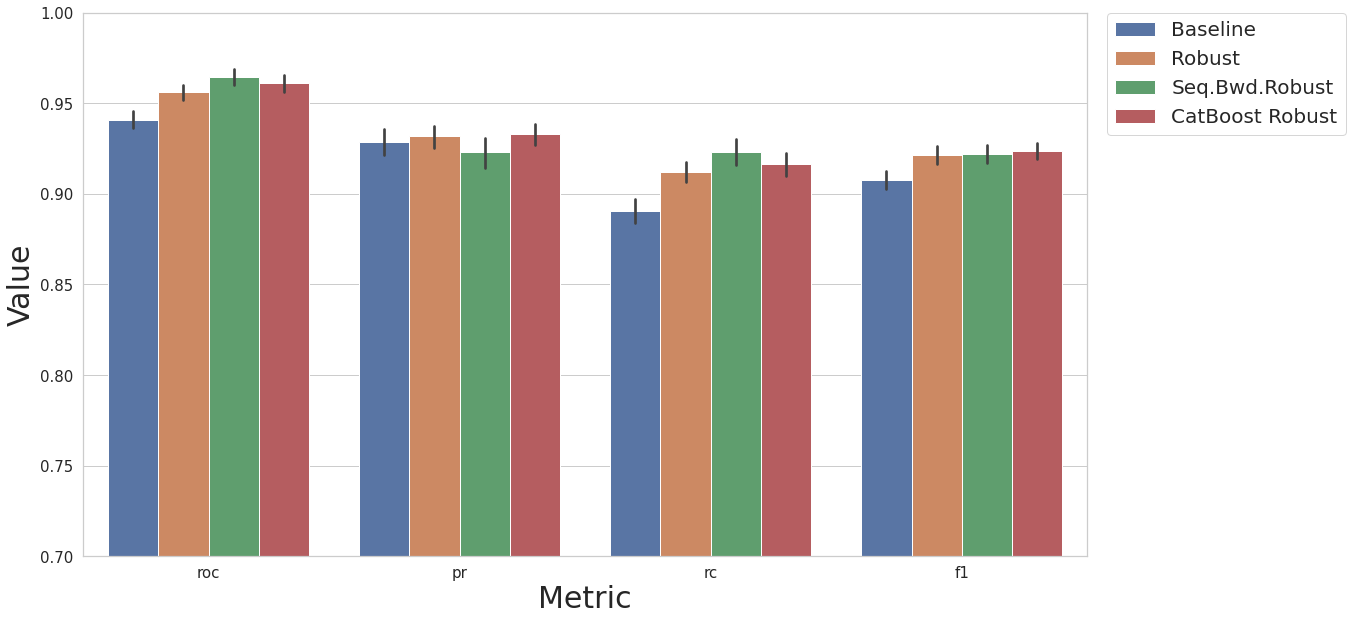
Feature Selection
Now we will perform feature selection, where all features are removed from the dataset to improve the model performance. We will compare two different feature selection pipelines with two baseline models.
Baseline KNN model
KNN model with robust scaler
KNN model, robust scaler, sequential feature selection
KNN model, robust scaler, CatBoost feature selection
[9]:
MODEL_NAME = "knn"
result_base = test_base(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(None, result_base, "Baseline")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataRobustScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Robust")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=False, scaler_ref=dp.DataRobustScaler, num_col=num_col, feat_sel_type="backward")
result_df = cs.add_results_df(result_df, restult_fs, "Seq.Bwd.Robust")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=False, scaler_ref=dp.DataRobustScaler, num_col=num_col, feat_sel_type='catboost')
result_df = cs.add_results_df(result_df, restult_fs, "CatBoost Robust")
[10]:
cs.plot_results(result_df, y_lim=[0.7, 1.0])
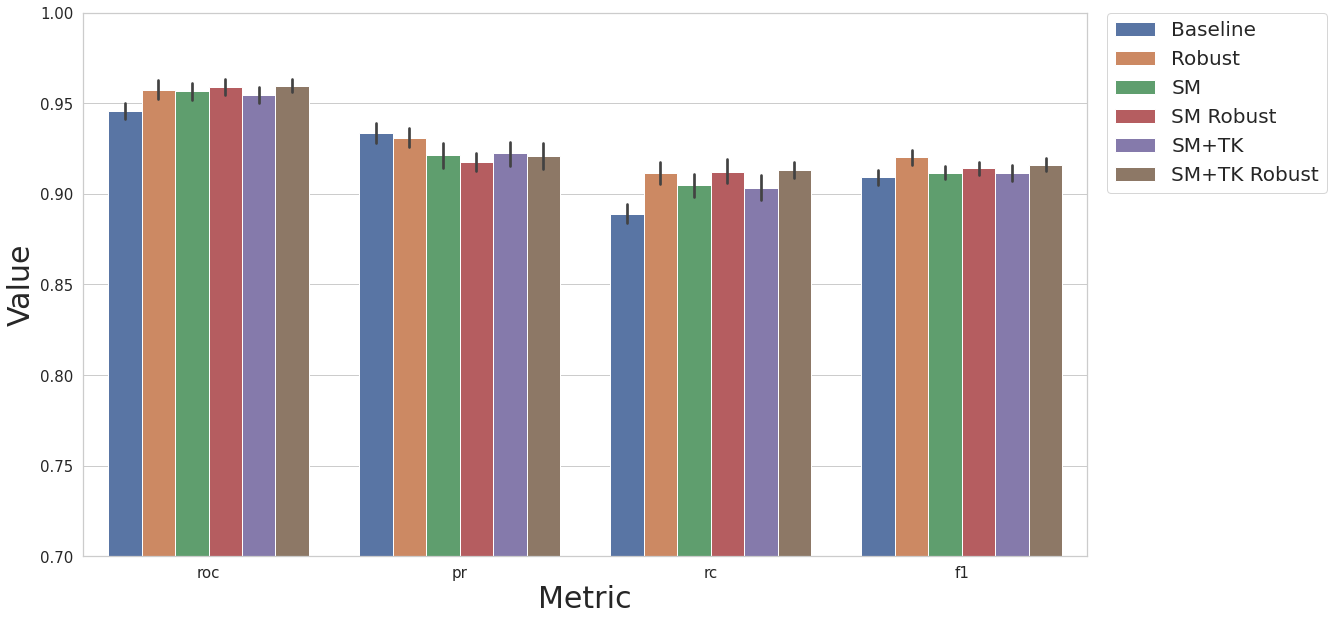
Artificial Instances - SMOTE
Since we have imbalanced classes (most trees are not diseased), we will experiment with creating artificial data.
baseline KNN model
baseline KNN model with robust scaler and removal of correlated features
KNN model, remove correlated features, SMOTE artificial data
KNN model, remove correlated features, robust scaler, SMOTE artificial data
KNN model, remove correlated features, SMOTE artificial data, TomekLink under sampling
KNN model, remove correlated features, robust scaler, SMOTE artificial data, TomekLink under sampling
[11]:
MODEL_NAME = "knn"
result_base = test_base(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(None, result_base, "Baseline")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataRobustScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Robust")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=None, feat_sel_type=None, art_str=0.2, under=False)
result_df = cs.add_results_df(result_df, restult_fs, "SM")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=dp.DataRobustScaler, num_col=num_col, feat_sel_type=None, art_str=0.2, under=False)
result_df = cs.add_results_df(result_df, restult_fs, "SM Robust")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=None, feat_sel_type=None, art_str=0.2, under=True)
result_df = cs.add_results_df(result_df, restult_fs, "SM+TK")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=dp.DataRobustScaler, num_col=num_col, feat_sel_type=None, art_str=0.2, under=True)
result_df = cs.add_results_df(result_df, restult_fs, "SM+TK Robust")
[12]:
cs.plot_results(result_df, y_lim=[0.7, 1.0])
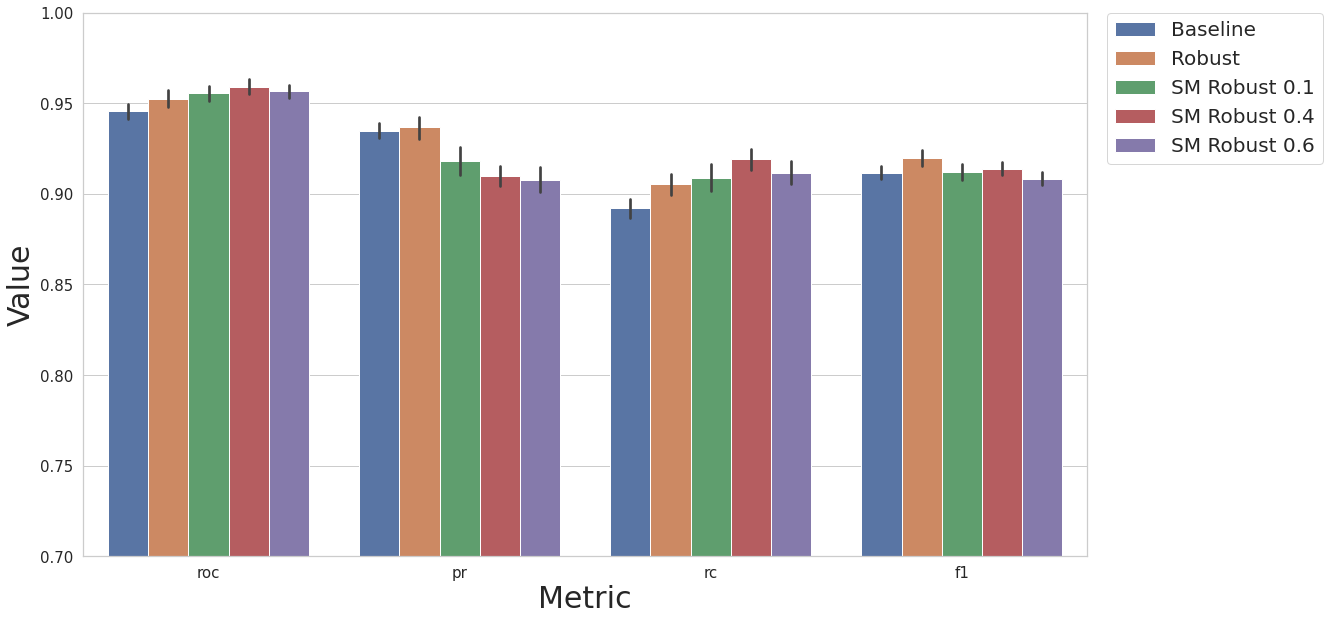
Now we will compare different quantities of artificial instances generated (the float refers to the ratio of minority class to majority class). All of these models contain correlated feature removal and the robust scaler.
Now we will compare different quantities of artificial instances generated (the float refers to the ratio of minority class to majority class). All of these models contain correlated feature removal and the robust scaler.
[13]:
MODEL_NAME = "knn"
result_base = test_base(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(None, result_base, "Baseline")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataRobustScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Robust")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=dp.DataRobustScaler, num_col=num_col, feat_sel_type=None, art_str=0.1, under=False)
result_df = cs.add_results_df(result_df, restult_fs, "SM Robust 0.1")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=dp.DataRobustScaler, num_col=num_col, feat_sel_type=None, art_str=0.4, under=False)
result_df = cs.add_results_df(result_df, restult_fs, "SM Robust 0.4")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=dp.DataRobustScaler, num_col=num_col, feat_sel_type=None, art_str=0.6, under=False)
result_df = cs.add_results_df(result_df, restult_fs, "SM Robust 0.6")
[14]:
cs.plot_results(result_df, y_lim=[0.7, 1.0])
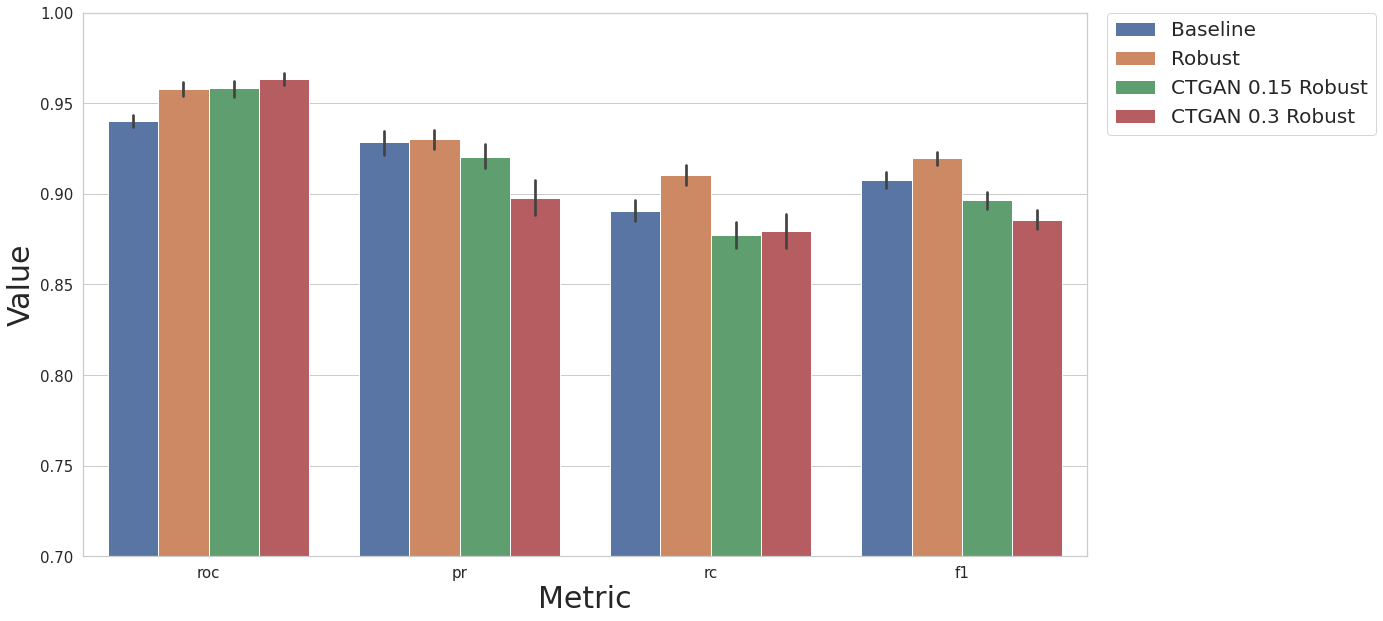
Artificial Instances - CTGAN
Instead of doing over sampling using SMOTE and its variations, we can create artificial instances of the minority class using the CTGAN. Here we perform the following experiments:
KNN baseline model
baseline KNN model with robust scaler and removal of correlated features
KNN model, robust scaler, removal of correlated features, over sampling with CTGAN ratio of 0.15
KNN model, robust scaler, removal of correlated features, over sampling with CTGAN ratio of 0.3
[15]:
MODEL_NAME = "knn"
result_base = test_base(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(None, result_base, "Baseline")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataRobustScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Robust")
restult_fs = test_ctgan_first(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=dp.DataRobustScaler, num_col=num_col, feat_sel_type=None, art_str=0.15, savefile="2_1.pkl")
result_df = cs.add_results_df(result_df, restult_fs, "CTGAN 0.15 Robust")
restult_fs = test_ctgan_first(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=dp.DataRobustScaler, num_col=num_col, feat_sel_type=None, art_str=0.3, savefile="2_2.pkl")
result_df = cs.add_results_df(result_df, restult_fs, "CTGAN 0.3 Robust")
[16]:
cs.plot_results(result_df, y_lim=[0.7, 1.0])
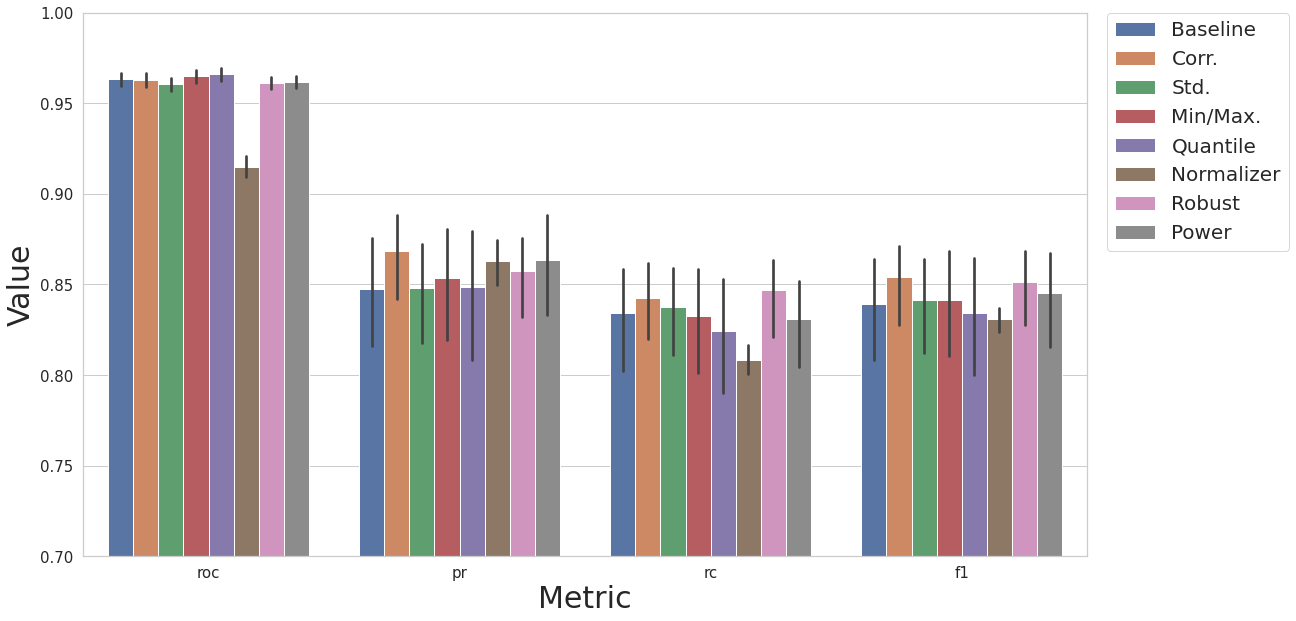
XGBoost
After the extensive experimenting with KNN models, we can also perform the same comparisons of experiments with XGBoost models instead. The following cells demonstrate this.
[17]:
MODEL_NAME = "xgb"
result_base = test_base(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(None, result_base, "Baseline")
result_cor = test_corr(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(result_df, result_cor, "Corr.")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataStandardScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Std.")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataMinMaxScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Min/Max.")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataQuantileTransformer, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Quantile")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataNormalizer, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Normalizer")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataRobustScaler, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Robust")
result_tr = test_corr_transf(df, label_col, N_EXEC, dp.DataPowerTransformer, MODEL_NAME, num_col)
result_df = cs.add_results_df(result_df, result_tr, "Power")
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
[18]:
cs.plot_results(result_df, y_lim=[0.7, 1.0])
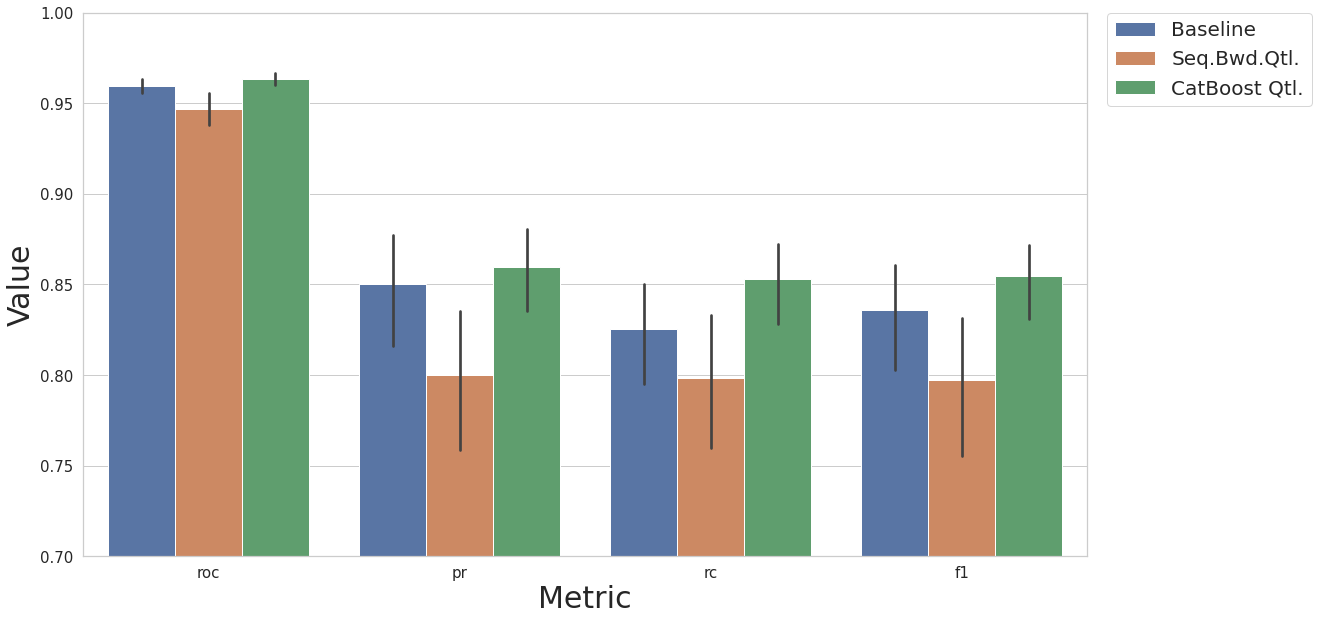
[19]:
result_base = test_base(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(None, result_base, "Baseline")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=None, feat_sel_type='backward')
result_df = cs.add_results_df(result_df, restult_fs, "Seq.Bwd.Qtl.")
restult_fs = test_smote_transf(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=None, feat_sel_type='catboost')
result_df = cs.add_results_df(result_df, restult_fs, "CatBoost Qtl.")
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
[20]:
cs.plot_results(result_df, y_lim=[0.7, 1.0])
[21]:
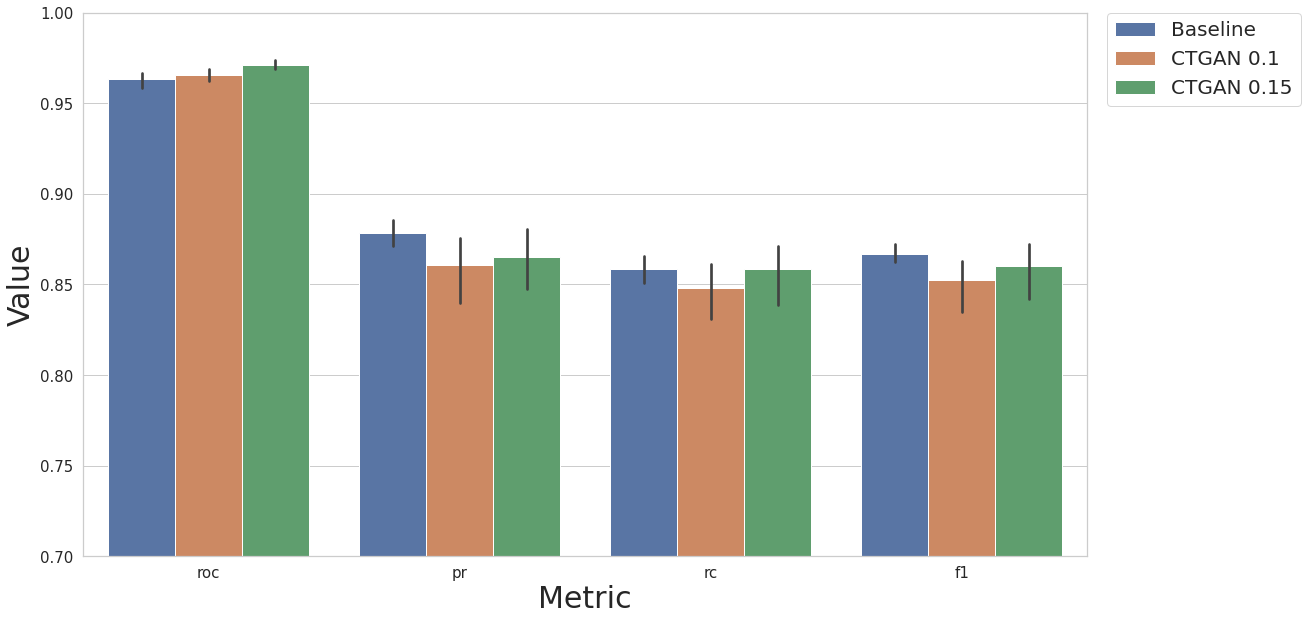
MODEL_NAME = "xgb"
result_base = test_base(df, label_col, N_EXEC, MODEL_NAME)
result_df = cs.add_results_df(None, result_base, "Baseline")
restult_fs = test_ctgan_first(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=None, feat_sel_type=None, art_str=0.1, savefile="2_3.pkl")
result_df = cs.add_results_df(result_df, restult_fs, "CTGAN 0.1")
restult_fs = test_ctgan_first(df, label_col, N_EXEC, MODEL_NAME, rcorr=True, scaler_ref=None, feat_sel_type=None, art_str=0.15, savefile="2_4.pkl")
result_df = cs.add_results_df(result_df, restult_fs, "CTGAN 0.15")
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
/home/mmendonca/ResponsibleAI/code/git/responsible-ai-mitigations/raimitigations/dataprocessing/model_utils.py:97: RuntimeWarning: invalid value encountered in true_divide
fscore = (2 * precision * recall) / (precision + recall)
[22]:
cs.plot_results(result_df, y_lim=[0.7, 1.0])