Summarise node centrality statistics with an igraph object
Source:R/network_summary.R
network_summary.RdPass an igraph object to the function and obtain centrality statistics for each node in the object as a data frame. This function works as a wrapper of the centralization functions in 'igraph'.
network_summary(graph, hrvar = NULL, return = "table")Arguments
- graph
'igraph' object that can be returned from
network_g2g()ornetwork_p2p()when thereturnargument is set to"network".- hrvar
String containing the name of the HR Variable by which to split metrics. Defaults to
NULL.- return
String specifying what output to return. Valid inputs include:
"table""network""plot"
See
Valuefor more information.
Value
By default, a data frame containing centrality statistics. Available statistics include:
betweenness: number of shortest paths going through a node.
closeness: number of steps required to access every other node from a given node.
degree: number of connections linked to a node.
eigenvector: a measure of the influence a node has on a network.
pagerank: calculates the PageRank for the specified vertices. Please refer to the igraph package documentation for the detailed technical definition.
When "network" is passed to "return", an 'igraph' object is returned with
additional node attributes containing centrality scores.
When "plot" is passed to "return", a summary table is returned showing
the average centrality scores by HR attribute. This is currently available if
there is a valid HR attribute.
See also
Examples
# Simulate a p2p network
p2p_data <- p2p_data_sim(size = 100)
g <- network_p2p(data = p2p_data, return = "network")
# Return summary table
network_summary(graph = g, return = "table")
#> # A tibble: 100 × 6
#> node_id betweenness closeness degree eigenvector pagerank
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 SIM_ID_1 0 0.339 9 0.652 0.00198
#> 2 SIM_ID_3 0 0.366 10 0.743 0.00198
#> 3 SIM_ID_4 66.8 0.337 11 0.816 0.00260
#> 4 SIM_ID_5 81.6 0.343 12 0.863 0.00261
#> 5 SIM_ID_6 66.8 0.295 12 0.877 0.00312
#> 6 SIM_ID_7 68.1 0.335 10 0.792 0.00277
#> 7 SIM_ID_8 49.4 0.304 12 0.892 0.00339
#> 8 SIM_ID_9 24.7 0.274 10 0.747 0.00363
#> 9 SIM_ID_10 49.4 0.289 12 0.853 0.00397
#> 10 SIM_ID_11 92.4 0.278 9 0.630 0.00387
#> # ℹ 90 more rows
# Return network with node centrality statistics
network_summary(graph = g, return = "network")
#> IGRAPH 491c1ee DNW- 100 500 --
#> + attr: weight (g/n), name (v/c), Organization (v/c), node_size (v/n),
#> | betweenness (v/n), closeness (v/n), degree (v/n), eigenvector (v/n),
#> | pagerank (v/n), weight (e/n)
#> + edges from 491c1ee (vertex names):
#> [1] SIM_ID_1->SIM_ID_4 SIM_ID_1->SIM_ID_5 SIM_ID_1->SIM_ID_6
#> [4] SIM_ID_1->SIM_ID_66 SIM_ID_1->SIM_ID_96 SIM_ID_1->SIM_ID_97
#> [7] SIM_ID_1->SIM_ID_98 SIM_ID_1->SIM_ID_2 SIM_ID_1->SIM_ID_100
#> [10] SIM_ID_3->SIM_ID_4 SIM_ID_3->SIM_ID_5 SIM_ID_3->SIM_ID_6
#> [13] SIM_ID_3->SIM_ID_7 SIM_ID_3->SIM_ID_8 SIM_ID_3->SIM_ID_32
#> [16] SIM_ID_3->SIM_ID_62 SIM_ID_3->SIM_ID_98 SIM_ID_3->SIM_ID_99
#> + ... omitted several edges
# Return summary plot
network_summary(graph = g, return = "plot", hrvar = "Organization")
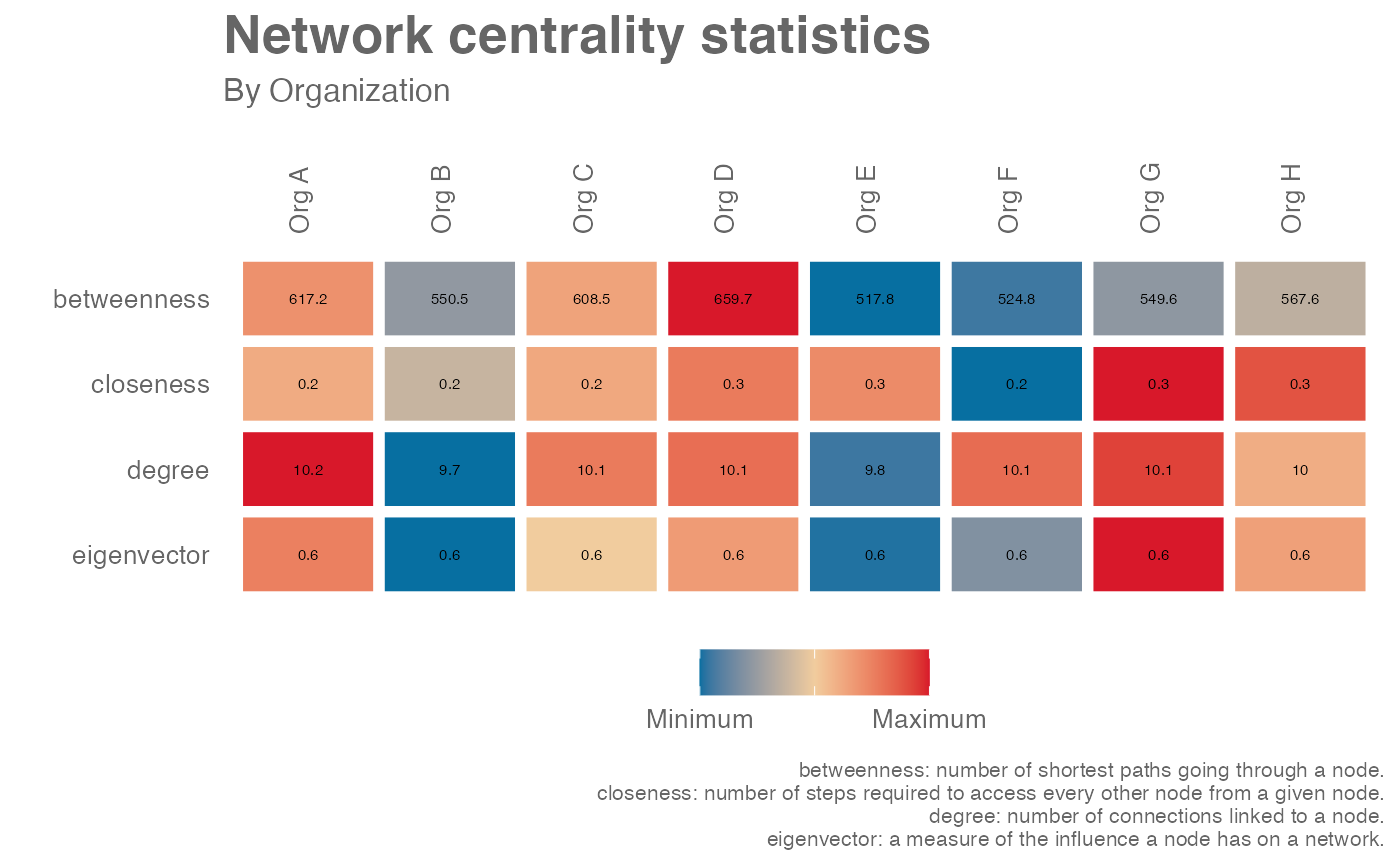 # Simulate a g2g network and return table
g2 <- g2g_data %>% network_g2g(return = "network")
#> `time_investor` field not provided. Assuming `TimeInvestors_Organization` as the `time_investor` variable.
#> `collaborator` field not provided. Assuming `Collaborators_Organization` as the `collaborator` variable.
network_summary(graph = g2, return = "table")
#> # A tibble: 6 × 6
#> node_id betweenness closeness degree eigenvector pagerank
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 "CEO" 0.488 1 5 0.581 0.126
#> 2 "Customer\nService" 0.364 0.714 7 0.829 0.166
#> 3 "Finance" 0.364 0.714 7 0.829 0.166
#> 4 "Financial\nPlanning" 0.182 0.714 6 0.662 0.147
#> 5 "Human\nResources" 1.02 0.833 8 0.873 0.188
#> 6 "IT" 1.59 0.833 9 1 0.208
# Simulate a g2g network and return table
g2 <- g2g_data %>% network_g2g(return = "network")
#> `time_investor` field not provided. Assuming `TimeInvestors_Organization` as the `time_investor` variable.
#> `collaborator` field not provided. Assuming `Collaborators_Organization` as the `collaborator` variable.
network_summary(graph = g2, return = "table")
#> # A tibble: 6 × 6
#> node_id betweenness closeness degree eigenvector pagerank
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 "CEO" 0.488 1 5 0.581 0.126
#> 2 "Customer\nService" 0.364 0.714 7 0.829 0.166
#> 3 "Finance" 0.364 0.714 7 0.829 0.166
#> 4 "Financial\nPlanning" 0.182 0.714 6 0.662 0.147
#> 5 "Human\nResources" 1.02 0.833 8 0.873 0.188
#> 6 "IT" 1.59 0.833 9 1 0.208