This page was generated from
docs/examples/DataSet/Accessing-data-in-DataSet.ipynb.
Interactive online version:
.
Accessing data in a DataSet¶
After a measurement is completed all the acquired data and metadata around it is accessible via a DataSet object. This notebook presents the useful methods and properties of the DataSet object which enable convenient access to the data, parameters information, and more. For general overview of the DataSet class, refer to DataSet class walkthrough.
Preparation: a DataSet from a dummy Measurement¶
In order to obtain a DataSet object, we are going to run a Measurement storing some dummy data (see notebook on Performing measurements using qcodes parameters and dataset notebook for more details).
[1]:
import os
import tempfile
import numpy as np
from qcodes.dataset import (
Measurement,
initialise_or_create_database_at,
load_or_create_experiment,
plot_dataset,
)
from qcodes.parameters import Parameter
[2]:
db_path = os.path.join(tempfile.gettempdir(), "data_access_example.db")
initialise_or_create_database_at(db_path)
exp = load_or_create_experiment(experiment_name="greco", sample_name="draco")
[3]:
x = Parameter(name="x", label="Voltage", unit="V", set_cmd=None, get_cmd=None)
t = Parameter(name="t", label="Time", unit="s", set_cmd=None, get_cmd=None)
y = Parameter(name="y", label="Voltage", unit="V", set_cmd=None, get_cmd=None)
y2 = Parameter(name="y2", label="Current", unit="A", set_cmd=None, get_cmd=None)
q = Parameter(name="q", label="Qredibility", unit="$", set_cmd=None, get_cmd=None)
[4]:
meas = Measurement(exp=exp, name="fresco")
meas.register_parameter(x)
meas.register_parameter(t)
meas.register_parameter(y, setpoints=(x, t))
meas.register_parameter(y2, setpoints=(x, t))
meas.register_parameter(q) # a standalone parameter
x_vals = np.linspace(-4, 5, 50)
t_vals = np.linspace(-500, 1500, 25)
with meas.run() as datasaver:
for xv in x_vals:
for tv in t_vals:
yv = np.sin(2 * np.pi * xv) * np.cos(2 * np.pi * 0.001 * tv) + 0.001 * tv
y2v = (
np.sin(2 * np.pi * xv) * np.cos(2 * np.pi * 0.001 * tv + 0.5 * np.pi)
- 0.001 * tv
)
datasaver.add_result((x, xv), (t, tv), (y, yv), (y2, y2v))
q_val = np.max(yv) - np.min(y2v) # a meaningless value
datasaver.add_result((q, q_val))
dataset = datasaver.dataset
Starting experimental run with id: 1.
For the sake of demonstrating what kind of data we’ve produced, let’s use plot_dataset to make some default plots of the data.
[5]:
plot_dataset(dataset)
[5]:
([<Axes: title={'center': 'Run #1, Experiment greco (draco)'}, xlabel='Voltage (V)', ylabel='Time (ks)'>,
<Axes: title={'center': 'Run #1, Experiment greco (draco)'}, xlabel='Voltage (V)', ylabel='Time (ks)'>],
[<matplotlib.colorbar.Colorbar at 0x7ff1561d1d50>,
<matplotlib.colorbar.Colorbar at 0x7ff156009210>])
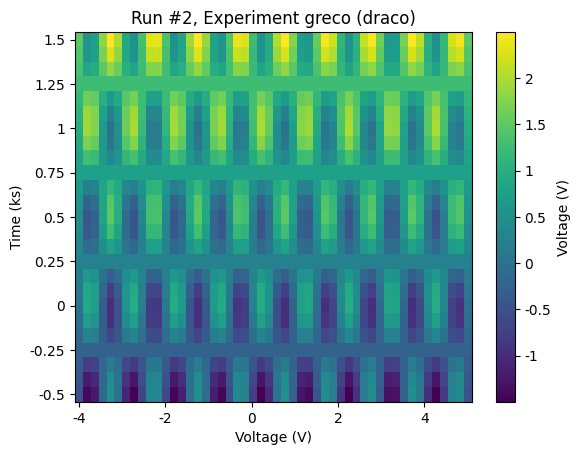
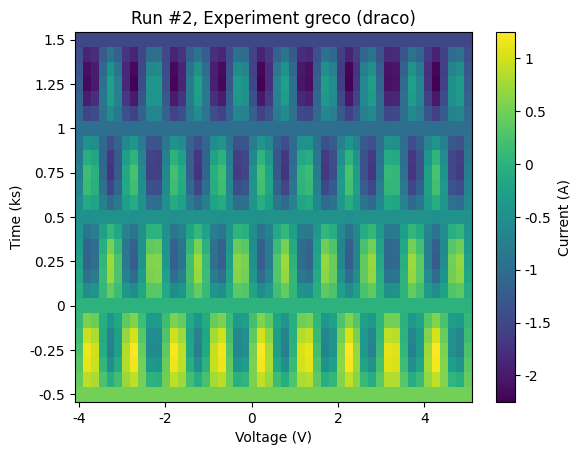
DataSet indentification¶
Before we dive into what’s in the DataSet, let’s briefly note how a DataSet is identified.
[6]:
dataset.captured_run_id
[6]:
1
[7]:
dataset.exp_name
[7]:
'greco'
[8]:
dataset.sample_name
[8]:
'draco'
[9]:
dataset.name
[9]:
'fresco'
Parameters in the DataSet¶
In this section we are getting information about the parameters stored in the given DataSet.
Why is that important? Let’s jump into data!
As it turns out, just “arrays of numbers” are not enough to reason about a given DataSet. Even comping up with a reasonable deafult plot, which is what plot_dataset does, requires information on DataSet’s parameters. In this notebook, we first have a detailed look at what is stored about parameters and how to work with this information. After that, we will cover data access methods.
Run description¶
Every dataset comes with a “description” (aka “run description”):
[10]:
dataset.description
[10]:
RunDescriber(InterDependencies_(dependencies={ParamSpecBase('y', 'numeric', 'Voltage', 'V'): (ParamSpecBase('x', 'numeric', 'Voltage', 'V'), ParamSpecBase('t', 'numeric', 'Time', 's')), ParamSpecBase('y2', 'numeric', 'Current', 'A'): (ParamSpecBase('x', 'numeric', 'Voltage', 'V'), ParamSpecBase('t', 'numeric', 'Time', 's'))}, inferences={}, standalones=frozenset({ParamSpecBase('q', 'numeric', 'Qredibility', '$')})), Shapes: None)
The description, an instance of RunDescriber object, is intended to describe the details of a dataset. In the future releases of QCoDeS it will likely be expanded. At the moment, it only contains an InterDependencies_ object under its interdeps attribute - which stores all the information about the parameters of the DataSet.
Let’s look into this InterDependencies_ object.
Interdependencies¶
Interdependencies_ object inside the run description contains information about all the parameters that are stored in the DataSet. Subsections below explain how the individual information about the parameters as well as their relationships are captured in the Interdependencies_ object.
[11]:
interdeps = dataset.description.interdeps
interdeps
[11]:
InterDependencies_(dependencies={ParamSpecBase('y', 'numeric', 'Voltage', 'V'): (ParamSpecBase('x', 'numeric', 'Voltage', 'V'), ParamSpecBase('t', 'numeric', 'Time', 's')), ParamSpecBase('y2', 'numeric', 'Current', 'A'): (ParamSpecBase('x', 'numeric', 'Voltage', 'V'), ParamSpecBase('t', 'numeric', 'Time', 's'))}, inferences={}, standalones=frozenset({ParamSpecBase('q', 'numeric', 'Qredibility', '$')}))
Dependencies, inferences, standalones¶
Information about every parameter is stored in the form of ParamSpecBase objects, and the releationship between parameters is captured via dependencies, inferences, and standalones attributes.
For example, the dataset that we are inspecting contains no inferences, and one standalone parameter q, and two dependent parameters y and y2, which both depend on independent x and t parameters:
[12]:
interdeps.inferences
[12]:
{}
[13]:
interdeps.standalones
[13]:
frozenset({ParamSpecBase('q', 'numeric', 'Qredibility', '$')})
[14]:
interdeps.dependencies
[14]:
{ParamSpecBase('y', 'numeric', 'Voltage', 'V'): (ParamSpecBase('x', 'numeric', 'Voltage', 'V'),
ParamSpecBase('t', 'numeric', 'Time', 's')),
ParamSpecBase('y2', 'numeric', 'Current', 'A'): (ParamSpecBase('x', 'numeric', 'Voltage', 'V'),
ParamSpecBase('t', 'numeric', 'Time', 's'))}
dependencies is a dictionary of ParamSpecBase objects. The keys are dependent parameters (those which depend on other parameters), and the corresponding values in the dictionary are tuples of independent parameters that the dependent parameter in the key depends on. Coloquially, each key-value pair of the dependencies dictionary is sometimes referred to as “parameter tree”.
inferences follows the same structure as dependencies.
standalones is a set - an unordered collection of ParamSpecBase objects representing “standalone” parameters, the ones which do not depend on other parameters, and no other parameter depends on them.
ParamSpecBase objects¶
ParamSpecBase object contains all the necessary information about a given parameter, for example, its name and unit:
[15]:
ps = next(iter(interdeps.dependencies.keys()))
print(f"Parameter {ps.name!r} is in {ps.unit!r}")
Parameter 'y' is in 'V'
paramspecs property returns a tuple of ParamSpecBases for all the parameters contained in the Interdependencies_ object:
[16]:
interdeps.paramspecs
[16]:
(ParamSpecBase('x', 'numeric', 'Voltage', 'V'),
ParamSpecBase('t', 'numeric', 'Time', 's'),
ParamSpecBase('y', 'numeric', 'Voltage', 'V'),
ParamSpecBase('y2', 'numeric', 'Current', 'A'),
ParamSpecBase('q', 'numeric', 'Qredibility', '$'))
Here’s a trivial example of iterating through dependent parameters of the Interdependencies_ object and extracting information about them from the ParamSpecBase objects:
[17]:
for d in interdeps.dependencies.keys():
print(f"Parameter {d.name!r} ({d.label}, {d.unit}) depends on:")
for i in interdeps.dependencies[d]:
print(f"- {i.name!r} ({i.label}, {i.unit})")
Parameter 'y' (Voltage, V) depends on:
- 'x' (Voltage, V)
- 't' (Time, s)
Parameter 'y2' (Current, A) depends on:
- 'x' (Voltage, V)
- 't' (Time, s)
Other useful methods and properties¶
Interdependencies_ object has a few useful properties and methods which make it easy to work it and with other Interdependencies_ and ParamSpecBase objects.
For example, top_level_parameters returns a tuple of all parameters that are not dependencies of other parameters:
[18]:
interdeps.top_level_parameters
[18]:
(ParamSpecBase('q', 'numeric', 'Qredibility', '$'),
ParamSpecBase('y', 'numeric', 'Voltage', 'V'),
ParamSpecBase('y2', 'numeric', 'Current', 'A'))
what_depends_on method allows to find what parameters depend on a given parameter:
[19]:
t_ps = interdeps.paramspecs[2]
t_deps = interdeps.what_depends_on(t_ps)
print(f"Following parameters depend on {t_ps.name!r} ({t_ps.label}, {t_ps.unit}):")
for t_dep in t_deps:
print(f"- {t_dep.name!r} ({t_dep.label}, {t_dep.unit})")
Following parameters depend on 'y' (Voltage, V):
Shortcuts to important parameters¶
For the frequently needed groups of parameters, DataSet object itself provides convenient methods and properties.
For example, use dependent_parameters property to get only dependent parameters of a given DataSet:
[20]:
dataset.dependent_parameters
[20]:
(ParamSpecBase('y', 'numeric', 'Voltage', 'V'),
ParamSpecBase('y2', 'numeric', 'Current', 'A'))
This is equivalent to:
[21]:
tuple(dataset.description.interdeps.dependencies.keys())
[21]:
(ParamSpecBase('y', 'numeric', 'Voltage', 'V'),
ParamSpecBase('y2', 'numeric', 'Current', 'A'))
Note on inferences¶
Inferences between parameters is a feature that has not been used yet within QCoDeS. The initial concepts around DataSet included it in order to link parameters that are not directly dependent on each other as “dependencies” are. It is very likely that “inferences” will be eventually deprecated and removed.
Note on ParamSpec’s¶
ParamSpecs originate from QCoDeS versions prior to0.2.0and for now are kept for backwards compatibility.ParamSpecs are completely superseded byInterDependencies_/ParamSpecBasebundle and will likely be deprecated in future versions of QCoDeS together with theDataSetmethods/properties that returnParamSpecs objects.
In addition to the Interdependencies_ object, DataSet also holds ParamSpec objects (not to be confused with ParamSpecBase objects from above). Similar to Interdependencies_ object, the ParamSpec objects hold information about parameters and their interdependencies but in a different way: for a given parameter, ParamSpec object itself contains information on names of parameters that it depends on, while for the InterDependencies_/ParamSpecBases this
information is stored only in the InterDependencies_ object.
DataSet exposes paramspecs property and get_parameters() method, both of which return ParamSpec objects of all the parameters of the dataset, and are not recommended for use:
[22]:
dataset.paramspecs
[22]:
{'x': ParamSpec('x', 'numeric', 'Voltage', 'V', inferred_from=[], depends_on=[]),
't': ParamSpec('t', 'numeric', 'Time', 's', inferred_from=[], depends_on=[]),
'y': ParamSpec('y', 'numeric', 'Voltage', 'V', inferred_from=[], depends_on=['x', 't']),
'y2': ParamSpec('y2', 'numeric', 'Current', 'A', inferred_from=[], depends_on=['x', 't']),
'q': ParamSpec('q', 'numeric', 'Qredibility', '$', inferred_from=[], depends_on=[])}
[23]:
dataset.get_parameters()
[23]:
[ParamSpec('x', 'numeric', 'Voltage', 'V', inferred_from=[], depends_on=[]),
ParamSpec('t', 'numeric', 'Time', 's', inferred_from=[], depends_on=[]),
ParamSpec('y', 'numeric', 'Voltage', 'V', inferred_from=[], depends_on=['x', 't']),
ParamSpec('y2', 'numeric', 'Current', 'A', inferred_from=[], depends_on=['x', 't']),
ParamSpec('q', 'numeric', 'Qredibility', '$', inferred_from=[], depends_on=[])]
[24]:
dataset.parameters
[24]:
'x,t,y,y2,q'
To give an example of what it takes to work with ParamSpec objects as opposed to Interdependencies_ object, here’s a function that one needs to write in order to find standalone ParamSpecs from a given list of ParamSpecs:
[25]:
def get_standalone_parameters(paramspecs):
all_independents = {spec.name for spec in paramspecs if len(spec.depends_on_) == 0}
used_independents = {d for spec in paramspecs for d in spec.depends_on_}
standalones = all_independents.difference(used_independents)
return tuple(ps for ps in paramspecs if ps.name in standalones)
all_parameters = dataset.get_parameters()
standalone_parameters = get_standalone_parameters(all_parameters)
standalone_parameters
[25]:
(ParamSpec('q', 'numeric', 'Qredibility', '$', inferred_from=[], depends_on=[]),)
Getting data from DataSet¶
In this section methods for retrieving the actual data from the DataSet are discussed.
get_parameter_data - the powerhorse¶
DataSet provides one main method of accessing data - get_parameter_data. It returns data for groups of dependent-parameter-and-its-independent-parameters in a form of a nested dictionary of numpy arrays:
[26]:
dataset.get_parameter_data()
[26]:
{'q': {'q': array([3. , 2.08558738, 2.259722 , 3.31510822, 3.99537911,
3.49071755, 2.40188947, 2.02507209, 2.80884137, 3.82017225,
3.85514276, 2.87212284, 2.04133215, 2.3517716 , 3.43388374,
3.99948622, 3.375267 , 2.30431745, 2.06153158, 2.93592978,
3.88659931, 3.78183148, 2.74634542, 2.01281822, 2.4544651 ,
3.5455349 , 3.98718178, 3.25365458, 2.21816852, 2.11340069,
3.06407022, 3.93846842, 3.69568255, 2.624733 , 2.00051378,
2.56611626, 3.6482284 , 3.95866785, 3.12787716, 2.14485724,
2.17982775, 3.19115863, 3.97492791, 3.59811053, 2.50928245,
2.00462089, 2.68489178, 3.740278 , 3.91441262, 3. ])},
'y': {'y': array([-0.5 , -0.41666667, -0.33333333, ..., 1.33333333,
1.41666667, 1.5 ], shape=(1250,)),
'x': array([-4., -4., -4., ..., 5., 5., 5.], shape=(1250,)),
't': array([-500. , -416.66666667, -333.33333333, ..., 1333.33333333,
1416.66666667, 1500. ], shape=(1250,))},
'y2': {'y2': array([ 0.5 , 0.41666667, 0.33333333, ..., -1.33333333,
-1.41666667, -1.5 ], shape=(1250,)),
'x': array([-4., -4., -4., ..., 5., 5., 5.], shape=(1250,)),
't': array([-500. , -416.66666667, -333.33333333, ..., 1333.33333333,
1416.66666667, 1500. ], shape=(1250,))}}
Avoid excessive calls to loading data¶
Note that this call actually reads the data of the DataSet and in case of a DataSet with a lot of data can take noticable amount of time. Hence, it is recommended to limit the number of times the same data gets loaded in order to speed up the user’s code.
Loading data of selected parameters¶
Sometimes data only for a particular parameter or parameters needs to be loaded. For example, let’s assume that after inspecting the InterDependencies_ object from dataset.description.interdeps, we concluded that we want to load data of the q parameter and the y2 parameter. In order to do that, we just pass the names of these parameters, or their ParamSpecBases to get_parameter_data call:
[27]:
q_param_spec = next(iter(interdeps.standalones))
q_param_spec
[27]:
ParamSpecBase('q', 'numeric', 'Qredibility', '$')
[28]:
y2_param_spec = interdeps.non_dependencies[-1]
y2_param_spec
/tmp/ipykernel_10252/2745425255.py:1: QCoDeSDeprecationWarning: non_dependencies returns incorrect results and is deprecated. Use top_level_parameters as an alternative.
y2_param_spec = interdeps.non_dependencies[-1]
[28]:
ParamSpecBase('y2', 'numeric', 'Current', 'A')
[29]:
dataset.get_parameter_data(q_param_spec, y2_param_spec)
[29]:
{'q': {'q': array([3. , 2.08558738, 2.259722 , 3.31510822, 3.99537911,
3.49071755, 2.40188947, 2.02507209, 2.80884137, 3.82017225,
3.85514276, 2.87212284, 2.04133215, 2.3517716 , 3.43388374,
3.99948622, 3.375267 , 2.30431745, 2.06153158, 2.93592978,
3.88659931, 3.78183148, 2.74634542, 2.01281822, 2.4544651 ,
3.5455349 , 3.98718178, 3.25365458, 2.21816852, 2.11340069,
3.06407022, 3.93846842, 3.69568255, 2.624733 , 2.00051378,
2.56611626, 3.6482284 , 3.95866785, 3.12787716, 2.14485724,
2.17982775, 3.19115863, 3.97492791, 3.59811053, 2.50928245,
2.00462089, 2.68489178, 3.740278 , 3.91441262, 3. ])},
'y2': {'y2': array([ 0.5 , 0.41666667, 0.33333333, ..., -1.33333333,
-1.41666667, -1.5 ], shape=(1250,)),
'x': array([-4., -4., -4., ..., 5., 5., 5.], shape=(1250,)),
't': array([-500. , -416.66666667, -333.33333333, ..., 1333.33333333,
1416.66666667, 1500. ], shape=(1250,))}}
to_pandas_dataframe_dict and to_pandas_dataframe - for pandas fans¶
DataSet provides two methods for accessing data with pandas - to_pandas_dataframe and to_pandas_dataframe_dict. The method to_pandas_dataframe_dict returns data for groups of dependent-parameter-and-its-independent-parameters in a form of a dictionary of pandas.DataFrame s, while to_pandas_dataframe returns a concatendated pandas.DataFrame for groups of dependent-parameter-and-its-independent-parameters:
[30]:
df_dict = dataset.to_pandas_dataframe_dict()
# For the sake of making this article more readable,
# we will print the contents of the `dfs` dictionary
# manually by calling `.head()` on each of the DataFrames
for parameter_name, df in df_dict.items():
print(f"DataFrame for parameter {parameter_name}")
print("-----------------------------")
print(f"{df.head()!r}")
print("")
DataFrame for parameter q
-----------------------------
q
0 3.000000
1 2.085587
2 2.259722
3 3.315108
4 3.995379
DataFrame for parameter y
-----------------------------
y
x t
-4.0 -500.000000 -0.500000
-416.666667 -0.416667
-333.333333 -0.333333
-250.000000 -0.250000
-166.666667 -0.166667
DataFrame for parameter y2
-----------------------------
y2
x t
-4.0 -500.000000 0.500000
-416.666667 0.416667
-333.333333 0.333333
-250.000000 0.250000
-166.666667 0.166667
Alternativly to concatinate the DataSet data into a single pandas Dataframe run the following:
[31]:
df = dataset.to_pandas_dataframe()
print(f"{df.head()!r}")
q y y2
0 3.000000 NaN NaN
1 2.085587 NaN NaN
2 2.259722 NaN NaN
3 3.315108 NaN NaN
4 3.995379 NaN NaN
/opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/qcodes/dataset/exporters/export_to_pandas.py:36: UserWarning: Independent parameter setpoints are not equal. Check concatenated output carefully. Please consider using `to_pandas_dataframe_dict` to export each independent parameter to its own dataframe.
warnings.warn(
Similar to get_parameter_data, to_pandas_dataframe_dict and to_pandas_dataframe_dict also supports retrieving data for a given parameter(s), as well as start/stop arguments.
Both to_pandas_dataframe and to_pandas_dataframe_dict is implemented based on get_parameter_data, hence the performance considerations mentioned above for get_parameter_data apply to these methods as well.
For more details on to_pandas_dataframe refer to Working with pandas and xarray article.
Exporting to other file formats¶
The dataset support exporting to netcdf and csv via the dataset.export method. See Exporting QCoDes Datasets for more information.
Data extraction into “other” formats¶
If the user desires to export a QCoDeS DataSet into a format that is not readily supported by DataSet methods, we recommend to use to_pandas_dataframe_dict or to_pandas_dataframe_dict first, and then convert the resulting DataFrame s into a the desired format. This is becuase pandas package already implements converting DataFrame to various popular formats including comma-separated text file (.csv), HDF (.hdf5), xarray, Excel (.xls, .xlsx), and more;
refer to Working with pandas and xarray article, and `pandas documentation <https://pandas.pydata.org/pandas-docs/stable/reference/frame.html#serialization-io-conversion>`__ for more information.
Refer to the docstrings of those methods for more information on how to use them.
[ ]: