This page was generated from
docs/examples/DataSet/Exporting-data-to-other-file-formats.ipynb.
Interactive online version:
.
Exporting QCoDes Datasets¶
This notebook demonstrates how we can export QCoDeS datasets to other file formats.
Setup¶
First, we borrow an example from the measurement notebook to have some data to work with.
[1]:
%matplotlib inline
import shutil
from pathlib import Path
import numpy as np
import qcodes as qc
from qcodes.dataset import (
Measurement,
initialise_or_create_database_at,
load_by_run_spec,
load_from_netcdf,
load_or_create_experiment,
plot_dataset,
)
from qcodes.instrument_drivers.mock_instruments import (
DummyInstrument,
DummyInstrumentWithMeasurement,
)
[2]:
# preparatory mocking of physical setup
dac = DummyInstrument("dac", gates=["ch1", "ch2"])
dmm = DummyInstrumentWithMeasurement("dmm", setter_instr=dac)
station = qc.Station(dmm, dac)
[3]:
initialise_or_create_database_at(
Path.cwd().parent / "example_output" / "export_example.db"
)
exp = load_or_create_experiment(
experiment_name="exporting_data", sample_name="no sample"
)
[4]:
meas = Measurement(exp)
meas.register_parameter(dac.ch1) # register the first independent parameter
meas.register_parameter(dac.ch2) # register the second independent parameter
meas.register_parameter(
dmm.v2, setpoints=(dac.ch1, dac.ch2)
) # register the dependent one
[4]:
<qcodes.dataset.measurements.Measurement at 0x7f14a1660190>
We then perform two very basic measurements using dummy instruments.
[5]:
# run a 2D sweep
with meas.run() as datasaver:
for v1 in np.linspace(-1, 0, 200, endpoint=False):
for v2 in np.linspace(-1, 1, 201):
dac.ch1(v1)
dac.ch2(v2)
val = dmm.v2.get()
datasaver.add_result((dac.ch1, v1), (dac.ch2, v2), (dmm.v2, val))
dataset1 = datasaver.dataset
Starting experimental run with id: 1.
[6]:
# run a 2D sweep
with meas.run() as datasaver:
for v1 in np.linspace(0, 1, 200, endpoint=False):
for v2 in np.linspace(1, 2, 201):
dac.ch1(v1)
dac.ch2(v2)
val = dmm.v2.get()
datasaver.add_result((dac.ch1, v1), (dac.ch2, v2), (dmm.v2, val))
dataset2 = datasaver.dataset
Starting experimental run with id: 2.
Exporting data manually¶
The dataset can be exported using the export method. Currently exporting to netcdf and csv is supported.
[7]:
dataset2.export("netcdf", path=Path.cwd().parent / "example_output")
The export_info attribute contains information about where the dataset has been exported to:
[8]:
dataset2.export_info
[8]:
ExportInfo(export_paths={'nc': '/home/runner/work/Qcodes/Qcodes/docs/examples/example_output/qcodes_2_dafb7712-0000-0000-0000-019c7059180d.nc'})
Looking at the signature of export we can see that in addition to the file format we can set the prefix and path to export to.
[9]:
?dataset2.export
Export data automatically¶
Datasets may also be exported automatically using the configuration options given in dataset config section. Here you can toggle if a dataset should be exported automatically using the export_automatic option as well as set the default type, prefix, elements in the name, and path. See the table here for the relevant configuration options.
For more information about how to configure QCoDeS datasets see the page about configuration in the QCoDeS docs.
By default datasets are exported into a folder next to the database with the same name but . replaced by _ e.g. if you store data to ~/experiments.db the exported files will be storred in ~/experiments_db This folder is automatically created if it does not exist.
Automatically post process exported datasets.¶
QCoDeD will attempt to call any EntryPoint registered for the group “qcodes.dataset.on_export”. This will allow a user to setup a function that can trigger post processing such as backup to cloud, external drive, plotting or post process analysis. Functions registered for this entry point group are expected to take a Path to the file as input and return None. Please consult the Setuptools
docs for more information on the use of EntryPoints. The entry point function must take the path to the exported file as a positional argument and take **kwargs for future compatibility. At the moment a single keyword argument automatic_export is passed to the function which indicates if the dataset was automatically or manually exported.
Importing exported datasets into a new database¶
The above dataset has been created in the following database
[10]:
qc.config.core.db_location
[10]:
'/home/runner/work/Qcodes/Qcodes/docs/examples/example_output/export_example.db'
Now lets imagine that we move the exported dataset to a different computer. To emulate this we will create a new database file and set it as the active database.
[11]:
initialise_or_create_database_at(
Path.cwd().parent / "example_output" / "reimport_example.db"
)
[12]:
qc.config.core.db_location
[12]:
'/home/runner/work/Qcodes/Qcodes/docs/examples/example_output/reimport_example.db'
We can then reload the dataset from the netcdf file as a DataSetInMem. This is a class that closely matches the regular DataSet class however its metadata may or may not be written to a database file and its data is not written to a database file. See more in In memory dataset . Concretely this means that the data captured in the dataset can be acceced via dataset.cache.data etc. and not via the methods directly on the dataset (dataset.get_parameter_data
…)
Note that it is currently only possible to reload a dataset from a netcdf export and not from a csv export. This is due to the fact that a csv export only contains the raw data and not the metadata needed to recreate a dataset.
[13]:
loaded_ds = load_from_netcdf(dataset2.export_info.export_paths["nc"])
[14]:
type(loaded_ds)
[14]:
qcodes.dataset.data_set_in_memory.DataSetInMem
However, we can still export the data to Pandasa and xarray.
[15]:
loaded_ds.cache.to_xarray_dataset()
[15]:
<xarray.Dataset> Size: 325kB
Dimensions: (dac_ch1: 200, dac_ch2: 201)
Coordinates:
* dac_ch1 (dac_ch1) float64 2kB 0.0 0.005 0.01 0.015 ... 0.985 0.99 0.995
* dac_ch2 (dac_ch2) float64 2kB 1.0 1.005 1.01 1.015 ... 1.985 1.99 1.995 2.0
Data variables:
dmm_v2 (dac_ch1, dac_ch2) float64 322kB 0.006531 0.006253 ... 0.0004438
Attributes: (12/15)
ds_name: results
sample_name: no sample
exp_name: exporting_data
snapshot: {"station": {"instruments": {"dmm": {"functions...
guid: dafb7712-0000-0000-0000-019c7059180d
run_timestamp: 2026-02-18 10:43:33
... ...
run_id: 2
run_description: {"version": 3, "interdependencies": {"paramspec...
parent_dataset_links: []
run_timestamp_raw: 1771411413.0111716
completed_timestamp_raw: 1771411417.8700118
export_info: {"export_paths": {"nc": "/home/runner/work/Qcod...And plot it using plot_dataset.
[16]:
plot_dataset(loaded_ds)
[16]:
([<Axes: title={'center': 'Run #2, Experiment exporting_data (no sample)'}, xlabel='Gate ch1 (mV)', ylabel='Gate ch2 (V)'>],
[<matplotlib.colorbar.Colorbar at 0x7f144d8ba110>])
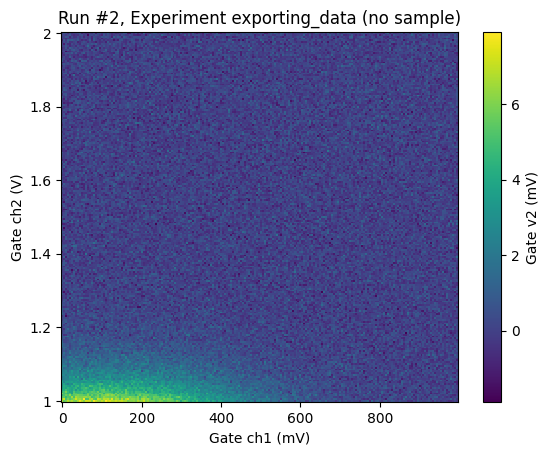
Note that the dataset will have the same captured_run_id and captured_counter as before:
[17]:
captured_run_id = loaded_ds.captured_run_id
captured_run_id
[17]:
2
But do note that the run_id and counter are in general not preserved since they represent the datasets number in a given db file.
[18]:
loaded_ds.run_id
[18]:
2
A loaded datasets metadata can be written to the current db file and subsequently the dataset including metadata and raw data reloaded from the database and netcdf file.
[19]:
loaded_ds.write_metadata_to_db()
Now that the metadata has been written to a database the dataset can be plotted with plottr like a regular dataset.
[20]:
del loaded_ds
[21]:
reloaded_ds = load_by_run_spec(captured_run_id=captured_run_id)
[22]:
plot_dataset(reloaded_ds)
[22]:
([<Axes: title={'center': 'Run #2, Experiment exporting_data (no sample)'}, xlabel='Gate ch1 (mV)', ylabel='Gate ch2 (V)'>],
[<matplotlib.colorbar.Colorbar at 0x7f14d8ded4d0>])
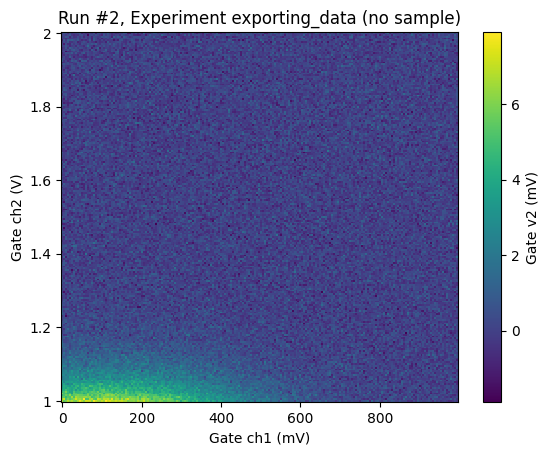
Note that loading a dataset from the database will also load the raw data into dataset.cache provided that the netcdf file is still in the location where file was when the metadata was written to the database. Load_by_runspec and related functions will load data into a regular DataSet provided that the data can be found in the database otherwise it will be loaded into a DataSetInMem
If the netcdf file cannot be found the dataset will load with a warning and the raw data will not be accessible from the dataset.
If this happens because you have moved the location of a netcdf file you can use the method set_netcdf_location to set a new location for the the netcdf file in the dataset and database file. Here we demonstrate this by copying the netcdf file and changing the location using this method.
[23]:
filepath = dataset2.export_info.export_paths["nc"]
new_file_path = str(Path.cwd().parent / "example_output" / "somefile.nc")
new_file_path
[23]:
'/home/runner/work/Qcodes/Qcodes/docs/examples/example_output/somefile.nc'
[24]:
shutil.copyfile(dataset2.export_info.export_paths["nc"], new_file_path)
[24]:
'/home/runner/work/Qcodes/Qcodes/docs/examples/example_output/somefile.nc'
[25]:
reloaded_ds.set_netcdf_location(new_file_path)
reloaded_ds.export_info
[25]:
ExportInfo(export_paths={'nc': '/home/runner/work/Qcodes/Qcodes/docs/examples/example_output/somefile.nc'})